Gene Page: POLR2A
Summary ?
| GeneID | 5430 |
| Symbol | POLR2A |
| Synonyms | POLR2|POLRA|RPB1|RPBh1|RPO2|RPOL2|RpIILS|hRPB220|hsRPB1 |
| Description | polymerase (RNA) II subunit A |
| Reference | MIM:180660|HGNC:HGNC:9187|Ensembl:ENSG00000181222|HPRD:08916|Vega:OTTHUMG00000177594 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.093 |
| Sherlock p-value | 0.992 |
| Fetal beta | -1.195 |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
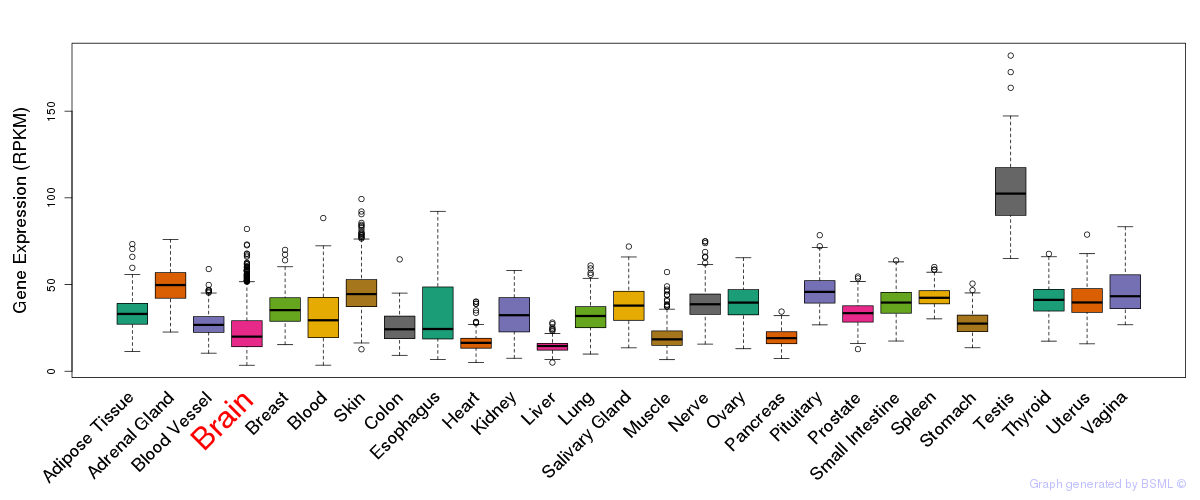
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHD3 | 0.74 | 0.67 |
| ZNF462 | 0.73 | 0.64 |
| BCL11A | 0.73 | 0.63 |
| UBTD2 | 0.72 | 0.67 |
| MYST4 | 0.72 | 0.64 |
| SMARCA4 | 0.72 | 0.67 |
| AUTS2 | 0.71 | 0.67 |
| ZNF860 | 0.71 | 0.60 |
| MLLT4 | 0.71 | 0.64 |
| GLB1L2 | 0.71 | 0.58 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.55 | -0.63 |
| C5orf53 | -0.54 | -0.65 |
| ALDOC | -0.54 | -0.64 |
| FBXO2 | -0.53 | -0.62 |
| PTH1R | -0.53 | -0.63 |
| CLU | -0.53 | -0.62 |
| TNFSF12 | -0.52 | -0.60 |
| AIFM3 | -0.52 | -0.64 |
| TSC22D4 | -0.52 | -0.63 |
| CSRP1 | -0.52 | -0.66 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | - | HPRD | 9710619 |
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD | 12154023 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 4506230 |9159119 |12154023|10725406 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 4506230 |9159119 |12154023 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-Western Co-purification | BioGRID | 9159119 |11504724 |12955082 |14506230 |
| C17orf56 | FLJ31528 | chromosome 17 open reading frame 56 | - | HPRD | 9710619 |
| CCNC | CycC | cyclin C | - | HPRD,BioGRID | 9443979 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Pol II interacts with cyclin D1 promoter. | BIND | 15808510 |
| CCNK | CPR4 | MGC9113 | cyclin K | Affinity Capture-Western in vitro | BioGRID | 9632813 |10574912 |
| CCNL2 | ANIA-6B | CCNM | DKFZp761A1210 | DKFZp762O195 | HCLA-ISO | HLA-ISO | PCEE | SB138 | cyclin L2 | - | HPRD,BioGRID | 14684736 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | POLR2A (pol II) interacts with the CD82 (KAI1) promoter. | BIND | 15829968 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | - | HPRD,BioGRID | 9443979 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | CDK9 interacts with and phosphorylates POLR2A (RNAPII). This interaction was modelled on a demonstrated interaction between human CDK9 and POLR2A from an unspecified species. | BIND | 12036313 |
| CIITA | C2TA | CIITAIV | MHC2TA | NLRA | class II, major histocompatibility complex, transactivator | Pol II interacts with p-MHC2TA. | BIND | 15502823 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 9710619 |
| CRKRS | CRK7 | CRKR | KIAA0904 | Cdc2-related kinase, arginine/serine-rich | - | HPRD | 11683387 |
| CSH2 | CS-2 | CSB | hCS-B | chorionic somatomammotropin hormone 2 | - | HPRD,BioGRID | 9312053 |10944529 |
| CTDP1 | CCFDN | FCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 9159119 |
| CTDSP1 | NLIIF | SCP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | Biochemical Activity | BioGRID | 12721286 |
| CTDSP2 | OS4 | PSR2 | SCP2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 | Biochemical Activity | BioGRID | 12721286 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD | 11149922 |
| DSCAM | CHD2-42 | CHD2-52 | Down syndrome cell adhesion molecule | Pol II interacts with DSCAM-1 enhancer. | BIND | 16009131 |
| EBAG9 | EB9 | PDAF | RCAS1 | estrogen receptor binding site associated, antigen, 9 | Pol II interacts with EBAG9 promoter. | BIND | 15808510 |
| ELP3 | FLJ10422 | KAT9 | elongation protein 3 homolog (S. cerevisiae) | Co-purification | BioGRID | 11714725 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western Co-purification | BioGRID | 9443979 |9710619 |
| FCP1 | FCP | FCPX | F-cell production 1 | Reconstituted Complex | BioGRID | 14576433 |
| GTF2B | TF2B | TFIIB | general transcription factor IIB | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | Co-purification Reconstituted Complex | BioGRID | 7926747 |9159119 |
| GTF2E2 | FE | TF2E2 | TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa | Affinity Capture-Western | BioGRID | 12665589 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Co-fractionation Co-purification Reconstituted Complex | BioGRID | 9159119 |9710619 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD,BioGRID | 3860504 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD | 9710619 |
| GTF2H4 | TFB2 | TFIIH | general transcription factor IIH, polypeptide 4, 52kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| HBB | CD113t-C | hemoglobin, beta | Beta-globin interacts with pol II. | BIND | 15565158 |
| HES1 | FLJ20408 | HES-1 | HHL | HRY | bHLHb39 | hairy and enhancer of split 1, (Drosophila) | HES1 promoter interacts with RNAPII. | BIND | 15546612 |
| HNRNPU | HNRPU | SAF-A | U21.1 | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | Affinity Capture-Western | BioGRID | 10490622 |
| HTATSF1 | TAT-SF1 | dJ196E23.2 | HIV-1 Tat specific factor 1 | Affinity Capture-Western | BioGRID | 10454543 |
| IFI6 | 6-16 | FAM14C | G1P3 | IFI-6-16 | IFI616 | interferon, alpha-inducible protein 6 | Pol II interacts with p-G1P3. | BIND | 15502823 |
| IL8 | CXCL8 | GCP-1 | GCP1 | LECT | LUCT | LYNAP | MDNCF | MONAP | NAF | NAP-1 | NAP1 | interleukin 8 | The IL-8 promoter interacts with Pol II. | BIND | 15494311 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Affinity Capture-Western | BioGRID | 9710619 |
| KLK2 | KLK2A2 | MGC12201 | hK2 | kallikrein-related peptidase 2 | Pol II interacts with p-KLK-2. | BIND | 15542435 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | Pol II interacts with p-PSA. | BIND | 15542435 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | Protein-peptide | BioGRID | 14578343 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | - | HPRD,BioGRID | 9710619 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Pol II interacts with c-myc promoter. | BIND | 15808510 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 12660246 |
| NONO | NMT55 | NRB54 | P54 | P54NRB | non-POU domain containing, octamer-binding | in vitro in vivo | BioGRID | 12358429 |
| NR0B2 | FLJ17090 | SHP | SHP1 | nuclear receptor subfamily 0, group B, member 2 | Reconstituted Complex | BioGRID | 12198243 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | Pol II interacts with NRIP-1 enhancer. | BIND | 16009131 |
| PCIF1 | C20orf67 | PDX1 C-terminal inhibiting factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12565871 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD | 10932246 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Affinity Capture-Western | BioGRID | 10393805 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | POLR2C (hRPB3) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2D | HSRBP4 | HSRPB4 | RBP4 | polymerase (RNA) II (DNA directed) polypeptide D | hRPB4 interacts with hRPB1. | BIND | 10567706 |
| POLR2E | RPABC1 | RPB5 | XAP4 | hRPB25 | hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | POLR2E (hRPB5) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2E | RPABC1 | RPB5 | XAP4 | hRPB25 | hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2F | HRBP14.4 | POLRF | RPABC2 | RPB14.4 | RPB6 | polymerase (RNA) II (DNA directed) polypeptide F | Reconstituted Complex | BioGRID | 9201987 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | Reconstituted Complex | BioGRID | 9201987 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | POLR2G (hRPB7) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | Reconstituted Complex | BioGRID | 9201987 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | POLR2H (hRPB8) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | hRPB1 interacts with hRPB8. | BIND | 10567706 |
| POLR2K | ABC10-alpha | RPABC4 | RPB10alpha | RPB12 | RPB7.0 | hRPB7.0 | hsRPB10a | polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | POLR2L (hRPB10-beta) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | Reconstituted Complex | BioGRID | 9201987 |
| PPIG | CARS-Cyp | CYP | MGC133241 | SRCyp | peptidylprolyl isomerase G (cyclophilin G) | - | HPRD,BioGRID | 9153302 |
| PPP1CC | PPP1G | protein phosphatase 1, catalytic subunit, gamma isoform | PPP1CC (TAPP) interacts with and dephosphorylates POLR2A (RNAPII). This interaction was modelled on a demonstrated interaction between human PPP1CC and POLR2A from an unspecified species. | BIND | 12036313 |
| RDBP | D6S45 | NELF-E | RD | RDP | RD RNA binding protein | Affinity Capture-Western | BioGRID | 12612062 |
| SAFB | DKFZp779C1727 | HAP | HET | SAFB1 | scaffold attachment factor B | - | HPRD,BioGRID | 9671816 |
| SMARCA2 | BAF190 | BRM | FLJ36757 | MGC74511 | SNF2 | SNF2L2 | SNF2LA | SWI2 | Sth1p | hBRM | hSNF2a | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | - | HPRD,BioGRID | 9710619 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-Western Co-purification in vitro | BioGRID | 8895581 |9443979 |9845365 |11238380 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD | 9443979 |12154023 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | - | HPRD,BioGRID | 9710619 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | - | HPRD | 9710619 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | - | HPRD | 9710619 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | - | HPRD | 9710619 |
| SMYD3 | FLJ21080 | MGC104324 | ZMYND1 | ZNFN3A1 | bA74P14.1 | SET and MYND domain containing 3 | Affinity Capture-Western | BioGRID | 15235609 |
| SND1 | TDRD11 | p100 | staphylococcal nuclease and tudor domain containing 1 | - | HPRD,BioGRID | 12234934 |
| SOD1 | ALS | ALS1 | IPOA | SOD | homodimer | superoxide dismutase 1, soluble | Pol II interacts with SOD-1 enhancer. | BIND | 16009131 |
| SUPT5H | FLJ34157 | SPT5 | SPT5H | suppressor of Ty 5 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 9450929 |10454543 |
| TAF10 | TAF2A | TAF2H | TAFII30 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | - | HPRD | 15099517 |
| TAF11 | MGC:15243 | PRO2134 | TAF2I | TAFII28 | TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Co-purification Reconstituted Complex | BioGRID | 9159119 |
| TCEA1 | GTF2S | SII | TCEA | TF2S | TFIIS | transcription elongation factor A (SII), 1 | - | HPRD,BioGRID | 12914699 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 9315662 |10908677 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | PolII interacts with TFF-1 enhancer. | BIND | 16009131 |
| TFF1 | BCEI | D21S21 | HP1.A | HPS2 | pNR-2 | pS2 | trefoil factor 1 | PolII interacts with TFF-1 promoter. | BIND | 16009131 |
| TRAK1 | OIP106 | trafficking protein, kinesin binding 1 | in vivo | BioGRID | 12435728 |
| TRAM2 | KIAA0057 | translocation associated membrane protein 2 | - | HPRD | 14612417 |
| UPF1 | FLJ43809 | FLJ46894 | HUPF1 | KIAA0221 | NORF1 | RENT1 | pNORF1 | UPF1 regulator of nonsense transcripts homolog (yeast) | - | HPRD | 6200476 |
| WHSC2 | FLJ10442 | FLJ25112 | NELF-A | NELFA | P/OKcl.15 | Wolf-Hirschhorn syndrome candidate 2 | Reconstituted Complex | BioGRID | 12612062 |
| XAB2 | DKFZp762C1015 | HCNP | HCRN | NTC90 | SYF1 | XPA binding protein 2 | - | HPRD,BioGRID | 10944529 |
| XBP1 | TREB5 | XBP2 | X-box binding protein 1 | PolII interacts with XBP-1 enhancer. | BIND | 16009131 |
| ZNF74 | Cos52 | ZNF520 | Zfp520 | zinc finger protein 74 | - | HPRD,BioGRID | 9346935 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG RNA POLYMERASE | 29 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RARRXR PATHWAY | 15 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME MICRORNA MIRNA BIOGENESIS | 23 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATORY RNA PATHWAYS | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA CAPPING | 30 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION COUPLED NER TC NER | 45 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE EXCISION REPAIR | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME ELONGATION ARREST AND RECOVERY | 32 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | 30 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | 23 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | 34 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN | 33 | 27 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES | 20 | 15 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |