Gene Page: POLR2H
Summary ?
| GeneID | 5437 |
| Symbol | POLR2H |
| Synonyms | RPABC3|RPB17|RPB8 |
| Description | polymerase (RNA) II subunit H |
| Reference | MIM:606023|HGNC:HGNC:9195|Ensembl:ENSG00000163882|HPRD:09082|Vega:OTTHUMG00000156746 |
| Gene type | protein-coding |
| Map location | 3q28 |
| Pascal p-value | 0.859 |
| Sherlock p-value | 0.731 |
| Fetal beta | -0.324 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16279464 | 3 | 184080807 | POLR2H | 1.37E-8 | -0.01 | 5.39E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
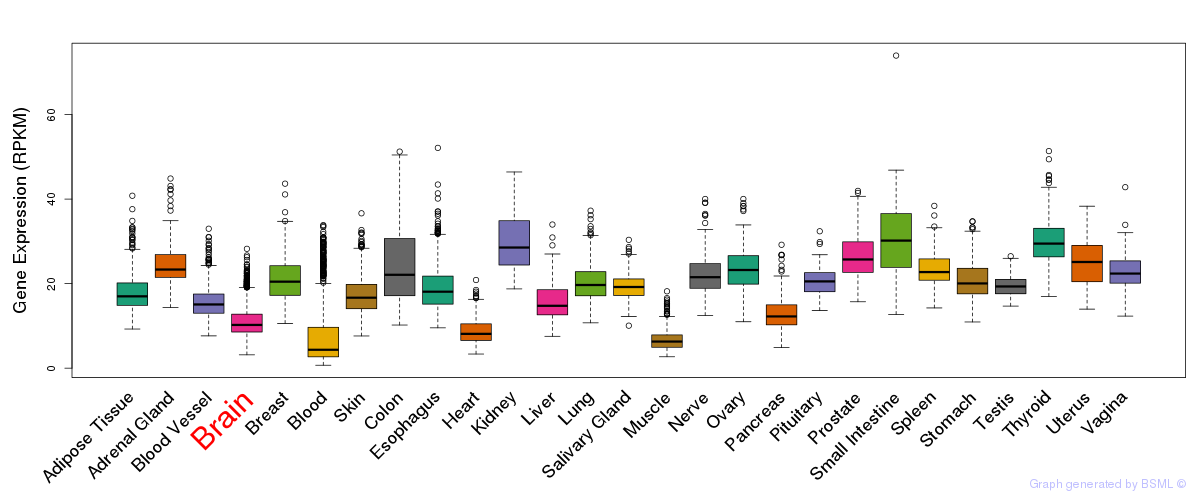
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CNOT6 | 0.98 | 0.97 |
| MYST3 | 0.98 | 0.96 |
| ARID2 | 0.97 | 0.98 |
| LATS1 | 0.97 | 0.98 |
| MSL2 | 0.97 | 0.97 |
| RFX7 | 0.97 | 0.98 |
| C11orf30 | 0.97 | 0.97 |
| CRKRS | 0.97 | 0.98 |
| PBRM1 | 0.97 | 0.98 |
| RBM16 | 0.97 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.73 | -0.89 |
| MT-CO2 | -0.72 | -0.90 |
| C5orf53 | -0.72 | -0.76 |
| FXYD1 | -0.72 | -0.89 |
| AIFM3 | -0.71 | -0.77 |
| AF347015.27 | -0.71 | -0.86 |
| HLA-F | -0.71 | -0.76 |
| IFI27 | -0.71 | -0.87 |
| S100B | -0.71 | -0.83 |
| AF347015.33 | -0.70 | -0.85 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | Affinity Capture-MS | BioGRID | 17353931 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS | BioGRID | 15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| NME2 | MGC111212 | NDPK-B | NDPKB | NM23-H2 | NM23B | puf | non-metastatic cells 2, protein (NM23B) expressed in | Affinity Capture-MS | BioGRID | 17353931 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | POLR2H (hRPB8) interacts with POLR2A (hRPB1). | BIND | 9201987 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | hRPB1 interacts with hRPB8. | BIND | 10567706 |
| POLR2B | POL2RB | RPB2 | hRPB140 | hsRPB2 | polymerase (RNA) II (DNA directed) polypeptide B, 140kDa | POLR2H (hRPB8) interacts weakly with POLR2B (hRPB2). | BIND | 9201987 |
| POLR2B | POL2RB | RPB2 | hRPB140 | hsRPB2 | polymerase (RNA) II (DNA directed) polypeptide B, 140kDa | hRPB2 interacts weakly with hRPB8. | BIND | 10567706 |
| POLR2B | POL2RB | RPB2 | hRPB140 | hsRPB2 | polymerase (RNA) II (DNA directed) polypeptide B, 140kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | POLR2H (hRPB8) interacts with POLR2C (hRPB3). | BIND | 9201987 |
| POLR2C | RPB3 | RPB31 | hRPB33 | hsRPB3 | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2D | HSRBP4 | HSRPB4 | RBP4 | polymerase (RNA) II (DNA directed) polypeptide D | hRPB4 interacts weakly with hRPB8. | BIND | 10567706 |
| POLR2E | RPABC1 | RPB5 | XAP4 | hRPB25 | hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | POLR2H (hRPB8) interacts with POLR2E (hRPB5). | BIND | 9201987 |
| POLR2E | RPABC1 | RPB5 | XAP4 | hRPB25 | hsRPB5 | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | POLR2H (hRPB8) interacts weakly with POLR2G (hRPB7). | BIND | 9201987 |
| POLR2G | MGC138367 | MGC138369 | RPB7 | hRPB19 | hsRPB7 | polymerase (RNA) II (DNA directed) polypeptide G | Reconstituted Complex | BioGRID | 9201987 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | POLR2H (hRPB8) interacts weakly with POLR2H (hRPB8). | BIND | 9201987 |
| POLR2H | RPABC3 | RPB17 | RPB8 | hsRPB8 | polymerase (RNA) II (DNA directed) polypeptide H | Reconstituted Complex | BioGRID | 9201987 |
| POLR2K | ABC10-alpha | RPABC4 | RPB10alpha | RPB12 | RPB7.0 | hRPB7.0 | hsRPB10a | polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa | POLR2H (hRPB8) interacts with POLR2K (hRPB10-alpha). | BIND | 9201987 |
| POLR2K | ABC10-alpha | RPABC4 | RPB10alpha | RPB12 | RPB7.0 | hRPB7.0 | hsRPB10a | polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | Reconstituted Complex | BioGRID | 9201987 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | hRPB8 interacts weakly with hRPB10-beta. | BIND | 10567706 |
| POLR3D | BN51T | RPC4 | RPC53 | TSBN51 | polymerase (RNA) III (DNA directed) polypeptide D, 44kDa | Affinity Capture-MS | BioGRID | 12391170 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG RNA POLYMERASE | 29 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | 23 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION TERMINATION | 22 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MICRORNA MIRNA BIOGENESIS | 23 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATORY RNA PATHWAYS | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA CAPPING | 30 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION COUPLED NER TC NER | 45 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE EXCISION REPAIR | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME ELONGATION ARREST AND RECOVERY | 32 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | 30 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | 169 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE | 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | 23 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION TERMINATION | 19 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | 34 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE | 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III CHAIN ELONGATION | 17 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| SLEBOS HEAD AND NECK CANCER WITH HPV UP | 84 | 43 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LUND SILENCED BY METHYLATION | 16 | 12 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| YAGI AML RELAPSE PROGNOSIS | 35 | 24 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| SCHUHMACHER MYC TARGETS UP | 80 | 57 | All SZGR 2.0 genes in this pathway |
| COLLER MYC TARGETS UP | 25 | 19 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| NADLER HYPERGLYCEMIA AT OBESITY | 58 | 35 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |