Gene Page: POMC
Summary ?
| GeneID | 5443 |
| Symbol | POMC |
| Synonyms | ACTH|CLIP|LPH|MSH|NPP|POC |
| Description | proopiomelanocortin |
| Reference | MIM:176830|HGNC:HGNC:9201|Ensembl:ENSG00000115138|HPRD:01462|Vega:OTTHUMG00000094764 |
| Gene type | protein-coding |
| Map location | 2p23.3 |
| Pascal p-value | 3.386E-5 |
| Sherlock p-value | 0.481 |
| Fetal beta | -0.585 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7565427 | chr2 | 25385637 | POMC | 5443 | 0.04 | cis | ||
| rs13031634 | chr2 | 25585652 | POMC | 5443 | 0.05 | cis | ||
| rs2197741 | chr3 | 108732108 | POMC | 5443 | 0.01 | trans | ||
| rs11733243 | chr4 | 4944450 | POMC | 5443 | 0.18 | trans | ||
| rs6063623 | chr20 | 49906075 | POMC | 5443 | 0.03 | trans | ||
| rs6067724 | chr20 | 49907840 | POMC | 5443 | 0.03 | trans | ||
| rs11696172 | chr20 | 49912433 | POMC | 5443 | 0.02 | trans | ||
| rs391008 | chrX | 44582706 | POMC | 5443 | 0.02 | trans |
Section II. Transcriptome annotation
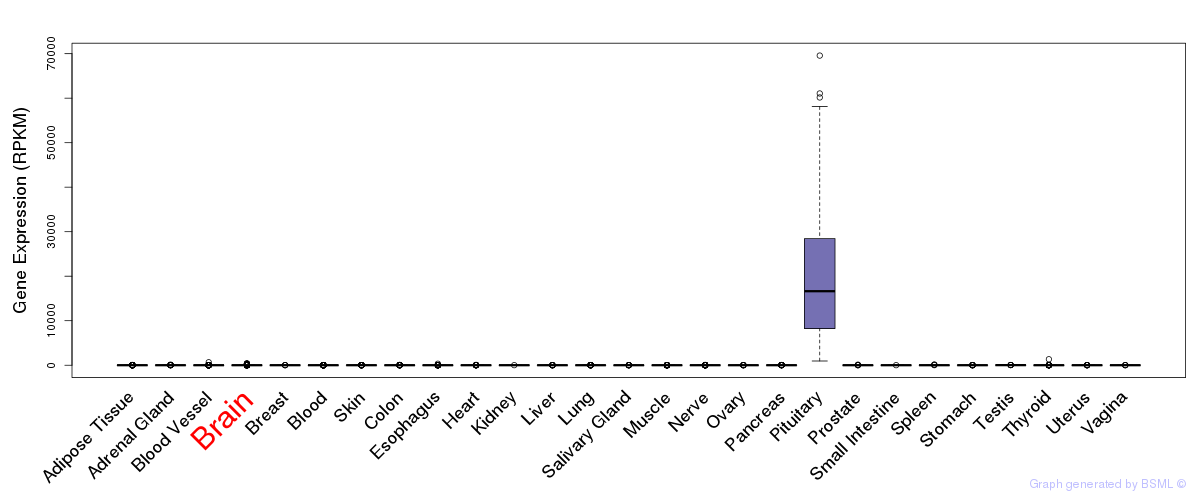
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM19A1 | 0.78 | 0.84 |
| RND1 | 0.72 | 0.77 |
| TCEAL1 | 0.72 | 0.80 |
| PRPS2 | 0.72 | 0.75 |
| AC012652.1 | 0.71 | 0.75 |
| PVRL3 | 0.69 | 0.74 |
| FABP3 | 0.68 | 0.75 |
| TM6SF1 | 0.67 | 0.79 |
| IL12RB2 | 0.67 | 0.77 |
| R3HDM1 | 0.67 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SORBS3 | -0.51 | -0.56 |
| PLXNB1 | -0.49 | -0.49 |
| CASKIN2 | -0.45 | -0.47 |
| NFKB2 | -0.42 | -0.51 |
| TRIP6 | -0.42 | -0.49 |
| EML3 | -0.41 | -0.43 |
| MAPKAPK3 | -0.41 | -0.45 |
| ACP6 | -0.41 | -0.43 |
| SLC9A3R1 | -0.40 | -0.43 |
| SLC2A4RG | -0.40 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005179 | hormone activity | IEA | - | |
| GO:0005179 | hormone activity | TAS | 9620771 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| GO:0008217 | regulation of blood pressure | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 9620771 | |
| GO:0006091 | generation of precursor metabolites and energy | TAS | 9620771 | |
| GO:0007165 | signal transduction | TAS | 9620771 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005625 | soluble fraction | TAS | 9620771 | |
| GO:0030141 | secretory granule | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | 35 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME ANDROGEN BIOSYNTHESIS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | 70 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOGENOUS STEROLS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE HORMONE BIOSYNTHESIS | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN ACTIVATION | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME STEROID HORMONES | 29 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 UP | 46 | 29 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR DN | 48 | 31 | All SZGR 2.0 genes in this pathway |
| MCGARVEY SILENCED BY METHYLATION IN COLON CANCER | 42 | 29 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |