Gene Page: GAR1
Summary ?
| GeneID | 54433 |
| Symbol | GAR1 |
| Synonyms | NOLA1 |
| Description | GAR1 ribonucleoprotein |
| Reference | MIM:606468|HGNC:HGNC:14264|Ensembl:ENSG00000109534|HPRD:07569|Vega:OTTHUMG00000161108 |
| Gene type | protein-coding |
| Map location | 4q25 |
| Pascal p-value | 0.38 |
| Sherlock p-value | 0.095 |
| Fetal beta | 0.84 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellum Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17404403 | 4 | 110737017 | GAR1 | 1.75E-5 | -0.377 | 0.016 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | GAR1 | 54433 | 0.09 | trans | ||
| rs11930428 | 4 | 110739840 | GAR1 | ENSG00000109534.12 | 1.464E-7 | 0.01 | 3174 | gtex_brain_ba24 |
| rs71888097 | 4 | 110742136 | GAR1 | ENSG00000109534.12 | 1.33E-7 | 0.01 | 5470 | gtex_brain_ba24 |
| rs3188897 | 4 | 110745425 | GAR1 | ENSG00000109534.12 | 1.31E-7 | 0.01 | 8759 | gtex_brain_ba24 |
| rs10365 | 4 | 110745854 | GAR1 | ENSG00000109534.12 | 1.309E-7 | 0.01 | 9188 | gtex_brain_ba24 |
| rs13128301 | 4 | 110750992 | GAR1 | ENSG00000109534.12 | 1.151E-7 | 0.01 | 14326 | gtex_brain_ba24 |
| rs35646628 | 4 | 110751439 | GAR1 | ENSG00000109534.12 | 1.138E-7 | 0.01 | 14773 | gtex_brain_ba24 |
| rs397779561 | 4 | 110755000 | GAR1 | ENSG00000109534.12 | 3.06E-7 | 0.01 | 18334 | gtex_brain_ba24 |
| rs11937249 | 4 | 110755321 | GAR1 | ENSG00000109534.12 | 3.293E-7 | 0.01 | 18655 | gtex_brain_ba24 |
| rs13107617 | 4 | 110757803 | GAR1 | ENSG00000109534.12 | 3.292E-7 | 0.01 | 21137 | gtex_brain_ba24 |
| rs7664699 | 4 | 110760941 | GAR1 | ENSG00000109534.12 | 5.467E-7 | 0.01 | 24275 | gtex_brain_ba24 |
| rs7438763 | 4 | 110763413 | GAR1 | ENSG00000109534.12 | 6.2E-7 | 0.01 | 26747 | gtex_brain_ba24 |
| rs35386791 | 4 | 110764207 | GAR1 | ENSG00000109534.12 | 7.411E-7 | 0.01 | 27541 | gtex_brain_ba24 |
| rs1990250 | 4 | 110770124 | GAR1 | ENSG00000109534.12 | 6.208E-7 | 0.01 | 33458 | gtex_brain_ba24 |
| rs2347129 | 4 | 110771788 | GAR1 | ENSG00000109534.12 | 5.928E-7 | 0.01 | 35122 | gtex_brain_ba24 |
| rs4698797 | 4 | 110773067 | GAR1 | ENSG00000109534.12 | 1.632E-6 | 0.01 | 36401 | gtex_brain_ba24 |
| rs6533469 | 4 | 110777387 | GAR1 | ENSG00000109534.12 | 1.944E-6 | 0.01 | 40721 | gtex_brain_ba24 |
| rs7654660 | 4 | 110784379 | GAR1 | ENSG00000109534.12 | 1.632E-6 | 0.01 | 47713 | gtex_brain_ba24 |
| rs6827802 | 4 | 110788540 | GAR1 | ENSG00000109534.12 | 1.632E-6 | 0.01 | 51874 | gtex_brain_ba24 |
| rs764205 | 4 | 110790911 | GAR1 | ENSG00000109534.12 | 1.632E-6 | 0.01 | 54245 | gtex_brain_ba24 |
| rs2159167 | 4 | 110823453 | GAR1 | ENSG00000109534.12 | 1.001E-6 | 0.01 | 86787 | gtex_brain_ba24 |
| rs7438961 | 4 | 110668651 | GAR1 | ENSG00000109534.12 | 1.628E-6 | 0.03 | -68015 | gtex_brain_putamen_basal |
| rs4698784 | 4 | 110669261 | GAR1 | ENSG00000109534.12 | 1.639E-6 | 0.03 | -67405 | gtex_brain_putamen_basal |
| rs11943677 | 4 | 110672770 | GAR1 | ENSG00000109534.12 | 1.535E-6 | 0.03 | -63896 | gtex_brain_putamen_basal |
| rs4541508 | 4 | 110682487 | GAR1 | ENSG00000109534.12 | 2.047E-6 | 0.03 | -54179 | gtex_brain_putamen_basal |
| rs6533457 | 4 | 110689557 | GAR1 | ENSG00000109534.12 | 2.257E-6 | 0.03 | -47109 | gtex_brain_putamen_basal |
| rs10365 | 4 | 110745854 | GAR1 | ENSG00000109534.12 | 2.404E-6 | 0.03 | 9188 | gtex_brain_putamen_basal |
| rs397779561 | 4 | 110755000 | GAR1 | ENSG00000109534.12 | 1.687E-6 | 0.03 | 18334 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
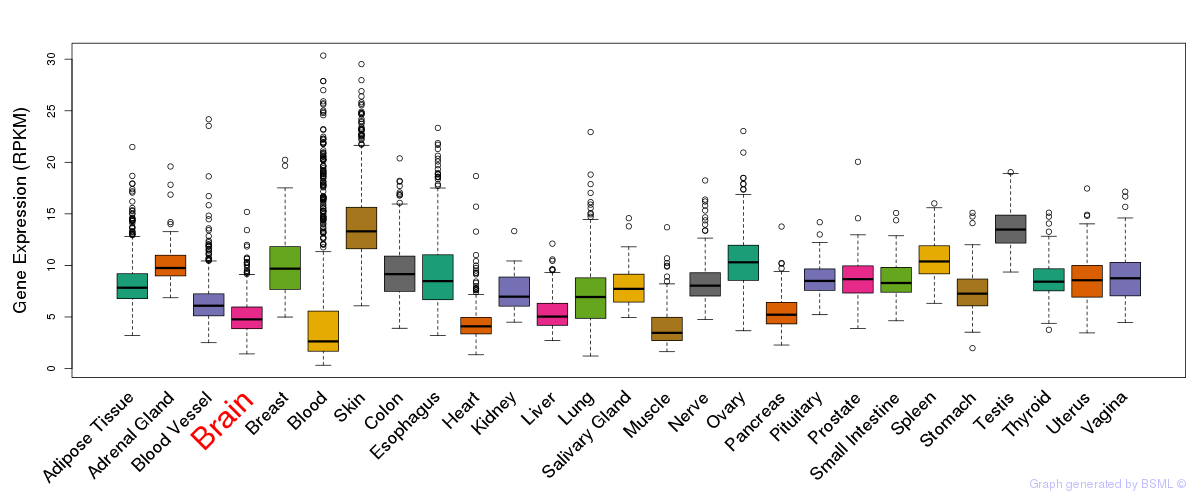
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 12P11 12 DN | 30 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |