Gene Page: POU3F2
Summary ?
| GeneID | 5454 |
| Symbol | POU3F2 |
| Synonyms | BRN2|N-Oct3|OCT7|OTF-7|OTF7|POUF3|brn-2|oct-7 |
| Description | POU class 3 homeobox 2 |
| Reference | MIM:600494|HGNC:HGNC:9215|Ensembl:ENSG00000184486|HPRD:02734|Vega:OTTHUMG00000015258 |
| Gene type | protein-coding |
| Map location | 6q16 |
| Pascal p-value | 0.152 |
| Fetal beta | 2.058 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04283429 | 6 | 99280108 | POU3F2 | 7.81E-9 | -0.03 | 3.77E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs920513 | chr1 | 120272605 | POU3F2 | 5454 | 0.15 | trans | ||
| rs10915848 | chr1 | 225788919 | POU3F2 | 5454 | 0.03 | trans | ||
| rs6544404 | chr2 | 41502544 | POU3F2 | 5454 | 0.14 | trans | ||
| rs1155107 | chr3 | 26843376 | POU3F2 | 5454 | 0 | trans | ||
| rs9848111 | chr3 | 26847784 | POU3F2 | 5454 | 0.2 | trans | ||
| rs1395163 | chr3 | 31028951 | POU3F2 | 5454 | 0.08 | trans | ||
| rs17029291 | chr3 | 32402138 | POU3F2 | 5454 | 1.095E-6 | trans | ||
| rs2197208 | chr3 | 105465403 | POU3F2 | 5454 | 0.2 | trans | ||
| rs1553189 | chr3 | 112939422 | POU3F2 | 5454 | 0 | trans | ||
| rs1807299 | chr3 | 134470799 | POU3F2 | 5454 | 0.04 | trans | ||
| rs6771817 | chr3 | 165557099 | POU3F2 | 5454 | 0.1 | trans | ||
| rs830202 | chr3 | 165654796 | POU3F2 | 5454 | 0.01 | trans | ||
| rs4645225 | chr4 | 126010830 | POU3F2 | 5454 | 0.05 | trans | ||
| rs750544 | chr5 | 51749054 | POU3F2 | 5454 | 8.811E-4 | trans | ||
| rs17146240 | chr5 | 119522933 | POU3F2 | 5454 | 0.01 | trans | ||
| rs17146261 | chr5 | 119534252 | POU3F2 | 5454 | 0.01 | trans | ||
| rs17165234 | chr5 | 126403634 | POU3F2 | 5454 | 0.06 | trans | ||
| rs2807510 | chr6 | 73090536 | POU3F2 | 5454 | 0.04 | trans | ||
| rs4299797 | chr6 | 86945707 | POU3F2 | 5454 | 0.19 | trans | ||
| rs9385758 | chr6 | 136729593 | POU3F2 | 5454 | 0.01 | trans | ||
| rs4617108 | chr7 | 49423831 | POU3F2 | 5454 | 0.03 | trans | ||
| rs3779536 | chr7 | 127233976 | POU3F2 | 5454 | 0 | trans | ||
| rs4615570 | chr8 | 13349013 | POU3F2 | 5454 | 0.02 | trans | ||
| rs1432030 | chr8 | 133175445 | POU3F2 | 5454 | 0.06 | trans | ||
| rs17060365 | chr9 | 77222282 | POU3F2 | 5454 | 0.14 | trans | ||
| rs7894597 | chr10 | 9384581 | POU3F2 | 5454 | 0.09 | trans | ||
| rs11000486 | chr10 | 74762547 | POU3F2 | 5454 | 0.17 | trans | ||
| rs12253630 | chr10 | 74768923 | POU3F2 | 5454 | 0.08 | trans | ||
| rs7078127 | chr10 | 74847633 | POU3F2 | 5454 | 0.04 | trans | ||
| rs12569493 | chr10 | 74886042 | POU3F2 | 5454 | 0 | trans | ||
| rs16930362 | chr10 | 74890320 | POU3F2 | 5454 | 4.305E-4 | trans | ||
| rs2271904 | chr10 | 74894374 | POU3F2 | 5454 | 0.04 | trans | ||
| rs7092445 | chr10 | 74921991 | POU3F2 | 5454 | 0.01 | trans | ||
| rs12256576 | chr10 | 74934313 | POU3F2 | 5454 | 0.1 | trans | ||
| rs1867565 | chr10 | 74977780 | POU3F2 | 5454 | 0.11 | trans | ||
| rs11000557 | chr10 | 74987393 | POU3F2 | 5454 | 0.06 | trans | ||
| rs16915029 | chr10 | 75001994 | POU3F2 | 5454 | 0.01 | trans | ||
| rs14488 | chr10 | 75007274 | POU3F2 | 5454 | 0.04 | trans | ||
| rs16930466 | chr10 | 75007587 | POU3F2 | 5454 | 0.01 | trans | ||
| rs11000579 | chr10 | 75027961 | POU3F2 | 5454 | 0.06 | trans | ||
| rs2183902 | chr10 | 75079279 | POU3F2 | 5454 | 0.13 | trans | ||
| rs11000607 | chr10 | 75089777 | POU3F2 | 5454 | 0 | trans | ||
| rs6480700 | chr10 | 75101689 | POU3F2 | 5454 | 0.03 | trans | ||
| rs16930573 | chr10 | 75167645 | POU3F2 | 5454 | 0 | trans | ||
| rs12571454 | chr10 | 75208954 | POU3F2 | 5454 | 0 | trans | ||
| rs17136959 | chr11 | 78373036 | POU3F2 | 5454 | 0.18 | trans | ||
| rs4944213 | chr11 | 78395188 | POU3F2 | 5454 | 0.18 | trans | ||
| rs17054422 | chr13 | 37317864 | POU3F2 | 5454 | 0.19 | trans | ||
| rs1610001 | chr13 | 100484419 | POU3F2 | 5454 | 0.02 | trans | ||
| rs1324669 | chr13 | 107872446 | POU3F2 | 5454 | 3.5E-8 | trans | ||
| rs1950367 | chr14 | 37657845 | POU3F2 | 5454 | 0.2 | trans | ||
| rs3742829 | chr14 | 73489267 | POU3F2 | 5454 | 0.03 | trans | ||
| rs8055506 | chr16 | 13661723 | POU3F2 | 5454 | 0 | trans | ||
| rs16962446 | chr16 | 13694936 | POU3F2 | 5454 | 0 | trans | ||
| rs17199331 | chr16 | 13747589 | POU3F2 | 5454 | 0.03 | trans | ||
| rs16958358 | chr17 | 55502410 | POU3F2 | 5454 | 4.672E-5 | trans | ||
| rs16949125 | chr18 | 45887469 | POU3F2 | 5454 | 0.19 | trans | ||
| rs1016438 | chr19 | 57591037 | POU3F2 | 5454 | 0.11 | trans | ||
| rs16987990 | chr20 | 41966000 | POU3F2 | 5454 | 0.05 | trans | ||
| rs6098023 | chr20 | 53113740 | POU3F2 | 5454 | 2.196E-4 | trans | ||
| rs6015314 | chr20 | 57167224 | POU3F2 | 5454 | 0 | trans | ||
| rs17145698 | chrX | 40218345 | POU3F2 | 5454 | 0 | trans | ||
| rs17328938 | chrX | 99519447 | POU3F2 | 5454 | 0.05 | trans | ||
| rs1335267 | chrX | 99570223 | POU3F2 | 5454 | 0.05 | trans | ||
| rs7065248 | chrX | 115510530 | POU3F2 | 5454 | 0.04 | trans |
Section II. Transcriptome annotation
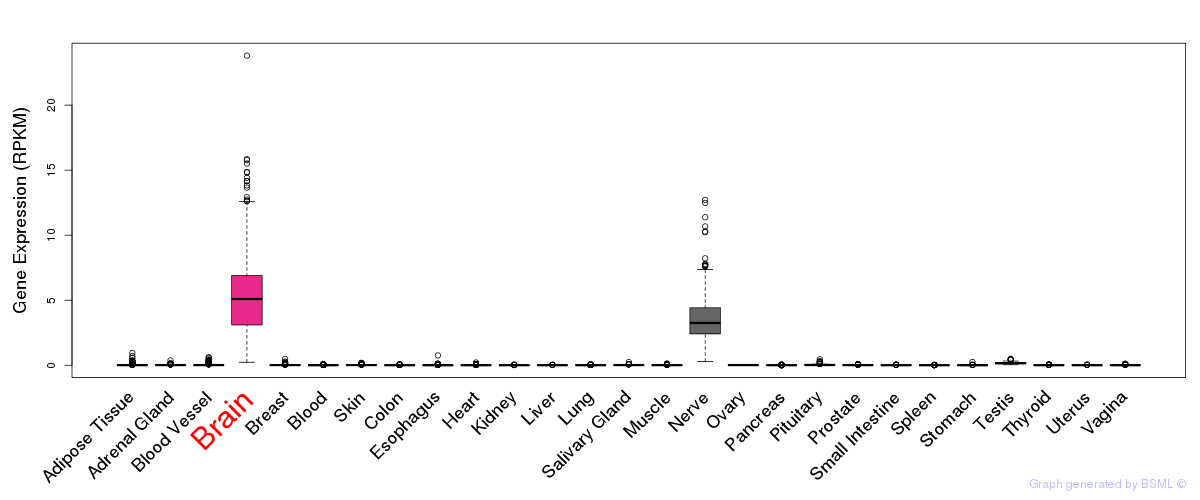
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 7601453 |9482665 | |
| GO:0042802 | identical protein binding | IPI | 11029584 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014002 | astrocyte development | IEA | neuron, astrocyte, Glial (GO term level: 10) | - |
| GO:0050770 | regulation of axonogenesis | IEA | neuron, axon (GO term level: 13) | - |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0008284 | positive regulation of cell proliferation | IDA | 15024079 | |
| GO:0008544 | epidermis development | IEA | - | |
| GO:0021985 | neurohypophysis development | IEA | - | |
| GO:0021979 | hypothalamus cell differentiation | IEA | - | |
| GO:0040018 | positive regulation of multicellular organism growth | IEA | - | |
| GO:0045595 | regulation of cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HBZ | 41 | 27 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER DN | 54 | 37 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER LATE RECURRENCE DN | 69 | 48 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 1646 | 1652 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 521 | 527 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-148/152 | 522 | 528 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-15/16/195/424/497 | 430 | 436 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-19 | 520 | 526 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-199 | 172 | 179 | 1A,m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-204/211 | 1695 | 1702 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-218 | 1678 | 1684 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-224 | 1923 | 1930 | 1A,m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-27 | 1646 | 1652 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-320 | 2404 | 2410 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-378 | 825 | 831 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU | ||||
| miR-410 | 2528 | 2534 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-494 | 1890 | 1896 | 1A | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU | ||||
| miR-495 | 1517 | 1523 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-542-3p | 1975 | 1981 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-544 | 2256 | 2263 | 1A,m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| miR-9 | 777 | 783 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.