Gene Page: TOMM7
Summary ?
| GeneID | 54543 |
| Symbol | TOMM7 |
| Synonyms | TOM7 |
| Description | translocase of outer mitochondrial membrane 7 |
| Reference | MIM:607980|HGNC:HGNC:21648|Ensembl:ENSG00000196683|HPRD:16266|Vega:OTTHUMG00000094805 |
| Gene type | protein-coding |
| Map location | 7p15.3 |
| Pascal p-value | 0.199 |
| Sherlock p-value | 0.924 |
| Fetal beta | -0.398 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26716323 | 7 | 22862328 | TOMM7 | -0.024 | 0.99 | DMG:Nishioka_2013 | |
| cg15703690 | 7 | 22766992 | TOMM7 | 0.007 | 4.414 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs57158381 | 7 | 22848987 | TOMM7 | ENSG00000196683.6 | 4.71E-7 | 0.01 | 13483 | gtex_brain_ba24 |
| rs2240728 | 7 | 22852499 | TOMM7 | ENSG00000196683.6 | 1.249E-7 | 0.01 | 9971 | gtex_brain_ba24 |
| rs2240727 | 7 | 22852512 | TOMM7 | ENSG00000196683.6 | 8.659E-7 | 0.01 | 9958 | gtex_brain_ba24 |
| rs1054471 | 7 | 22852643 | TOMM7 | ENSG00000196683.6 | 1.236E-7 | 0.01 | 9827 | gtex_brain_ba24 |
| rs2240726 | 7 | 22853135 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | 9335 | gtex_brain_ba24 |
| rs10950922 | 7 | 22854006 | TOMM7 | ENSG00000196683.6 | 8.984E-7 | 0.01 | 8464 | gtex_brain_ba24 |
| rs2286507 | 7 | 22854760 | TOMM7 | ENSG00000196683.6 | 8.791E-7 | 0.01 | 7710 | gtex_brain_ba24 |
| rs2286506 | 7 | 22854815 | TOMM7 | ENSG00000196683.6 | 8.659E-7 | 0.01 | 7655 | gtex_brain_ba24 |
| rs2286505 | 7 | 22854840 | TOMM7 | ENSG00000196683.6 | 8.777E-7 | 0.01 | 7630 | gtex_brain_ba24 |
| rs2286504 | 7 | 22854847 | TOMM7 | ENSG00000196683.6 | 8.667E-7 | 0.01 | 7623 | gtex_brain_ba24 |
| rs35416658 | 7 | 22855151 | TOMM7 | ENSG00000196683.6 | 8.717E-7 | 0.01 | 7319 | gtex_brain_ba24 |
| rs12113458 | 7 | 22855628 | TOMM7 | ENSG00000196683.6 | 8.659E-7 | 0.01 | 6842 | gtex_brain_ba24 |
| rs2286503 | 7 | 22856606 | TOMM7 | ENSG00000196683.6 | 8.411E-7 | 0.01 | 5864 | gtex_brain_ba24 |
| rs2286501 | 7 | 22856785 | TOMM7 | ENSG00000196683.6 | 8.373E-7 | 0.01 | 5685 | gtex_brain_ba24 |
| rs2286499 | 7 | 22857477 | TOMM7 | ENSG00000196683.6 | 8.261E-7 | 0.01 | 4993 | gtex_brain_ba24 |
| rs2286498 | 7 | 22857504 | TOMM7 | ENSG00000196683.6 | 8.261E-7 | 0.01 | 4966 | gtex_brain_ba24 |
| rs6461669 | 7 | 22858066 | TOMM7 | ENSG00000196683.6 | 8.667E-7 | 0.01 | 4404 | gtex_brain_ba24 |
| rs6949233 | 7 | 22858191 | TOMM7 | ENSG00000196683.6 | 8.176E-7 | 0.01 | 4279 | gtex_brain_ba24 |
| rs10259561 | 7 | 22859093 | TOMM7 | ENSG00000196683.6 | 7.088E-7 | 0.01 | 3377 | gtex_brain_ba24 |
| rs4722188 | 7 | 22860211 | TOMM7 | ENSG00000196683.6 | 1.182E-7 | 0.01 | 2259 | gtex_brain_ba24 |
| rs4722189 | 7 | 22860474 | TOMM7 | ENSG00000196683.6 | 1.18E-7 | 0.01 | 1996 | gtex_brain_ba24 |
| rs4719716 | 7 | 22861369 | TOMM7 | ENSG00000196683.6 | 1.286E-7 | 0.01 | 1101 | gtex_brain_ba24 |
| rs981792 | 7 | 22861639 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | 831 | gtex_brain_ba24 |
| rs2270108 | 7 | 22862131 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | 339 | gtex_brain_ba24 |
| rs2270107 | 7 | 22862183 | TOMM7 | ENSG00000196683.6 | 9.337E-8 | 0.01 | 287 | gtex_brain_ba24 |
| rs2270105 | 7 | 22862467 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | 3 | gtex_brain_ba24 |
| rs2270103 | 7 | 22862722 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | -252 | gtex_brain_ba24 |
| rs10276099 | 7 | 22862950 | TOMM7 | ENSG00000196683.6 | 1.19E-7 | 0.01 | -480 | gtex_brain_ba24 |
| rs1304431 | 7 | 22865412 | TOMM7 | ENSG00000196683.6 | 8.659E-7 | 0.01 | -2942 | gtex_brain_ba24 |
| rs1029740 | 7 | 22873586 | TOMM7 | ENSG00000196683.6 | 8.042E-7 | 0.01 | -11116 | gtex_brain_ba24 |
| rs929464 | 7 | 22874770 | TOMM7 | ENSG00000196683.6 | 8.659E-7 | 0.01 | -12300 | gtex_brain_ba24 |
| rs17724311 | 7 | 22876793 | TOMM7 | ENSG00000196683.6 | 2.121E-6 | 0.01 | -14323 | gtex_brain_ba24 |
| rs17147344 | 7 | 22880856 | TOMM7 | ENSG00000196683.6 | 2.172E-6 | 0.01 | -18386 | gtex_brain_ba24 |
| rs57158381 | 7 | 22848987 | TOMM7 | ENSG00000196683.6 | 2.749E-7 | 0 | 13483 | gtex_brain_putamen_basal |
| rs2240728 | 7 | 22852499 | TOMM7 | ENSG00000196683.6 | 4.045E-7 | 0 | 9971 | gtex_brain_putamen_basal |
| rs1054471 | 7 | 22852643 | TOMM7 | ENSG00000196683.6 | 4.071E-7 | 0 | 9827 | gtex_brain_putamen_basal |
| rs2240726 | 7 | 22853135 | TOMM7 | ENSG00000196683.6 | 4.173E-7 | 0 | 9335 | gtex_brain_putamen_basal |
| rs4722188 | 7 | 22860211 | TOMM7 | ENSG00000196683.6 | 1.679E-7 | 0 | 2259 | gtex_brain_putamen_basal |
| rs4722189 | 7 | 22860474 | TOMM7 | ENSG00000196683.6 | 1.579E-7 | 0 | 1996 | gtex_brain_putamen_basal |
| rs4719716 | 7 | 22861369 | TOMM7 | ENSG00000196683.6 | 1.119E-7 | 0 | 1101 | gtex_brain_putamen_basal |
| rs981792 | 7 | 22861639 | TOMM7 | ENSG00000196683.6 | 1.572E-7 | 0 | 831 | gtex_brain_putamen_basal |
| rs2270108 | 7 | 22862131 | TOMM7 | ENSG00000196683.6 | 1.572E-7 | 0 | 339 | gtex_brain_putamen_basal |
| rs2270107 | 7 | 22862183 | TOMM7 | ENSG00000196683.6 | 2.651E-7 | 0 | 287 | gtex_brain_putamen_basal |
| rs2270105 | 7 | 22862467 | TOMM7 | ENSG00000196683.6 | 1.572E-7 | 0 | 3 | gtex_brain_putamen_basal |
| rs2270103 | 7 | 22862722 | TOMM7 | ENSG00000196683.6 | 1.572E-7 | 0 | -252 | gtex_brain_putamen_basal |
| rs10276099 | 7 | 22862950 | TOMM7 | ENSG00000196683.6 | 1.572E-7 | 0 | -480 | gtex_brain_putamen_basal |
| rs17147344 | 7 | 22880856 | TOMM7 | ENSG00000196683.6 | 4.636E-7 | 0 | -18386 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
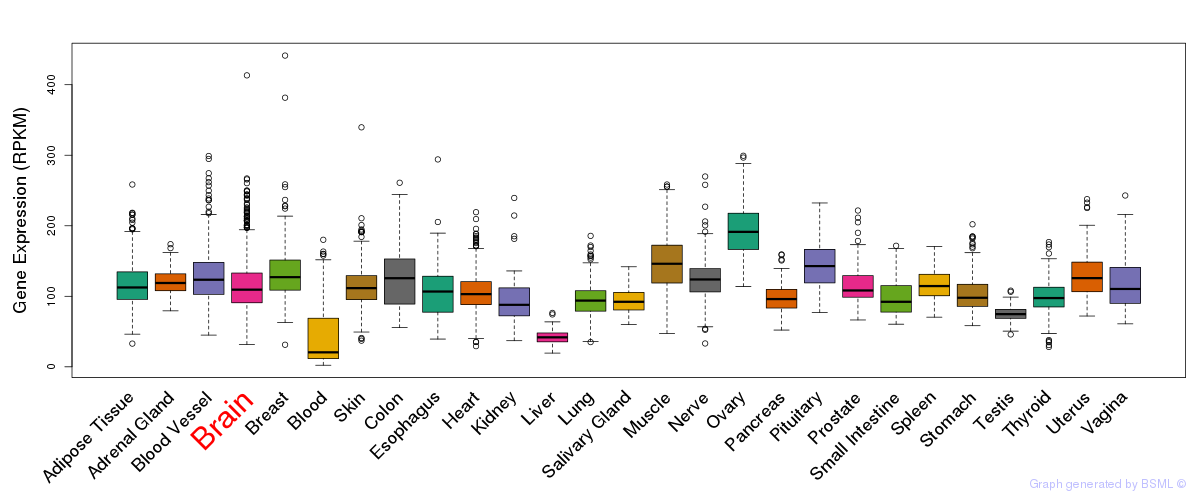
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME MITOCHONDRIAL PROTEIN IMPORT | 58 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |