Gene Page: NECAB2
Summary ?
| GeneID | 54550 |
| Symbol | NECAB2 |
| Synonyms | EFCBP2 |
| Description | N-terminal EF-hand calcium binding protein 2 |
| Reference | HGNC:HGNC:23746|Ensembl:ENSG00000103154|HPRD:13263|Vega:OTTHUMG00000137636 |
| Gene type | protein-coding |
| Map location | 16q23.3 |
| Pascal p-value | 0.248 |
| Sherlock p-value | 0.894 |
| Fetal beta | -2.481 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00021600 | 16 | 84003607 | NECAB2 | 2.806E-4 | 0.856 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17542055 | chr7 | 42652803 | NECAB2 | 54550 | 0.15 | trans |
Section II. Transcriptome annotation
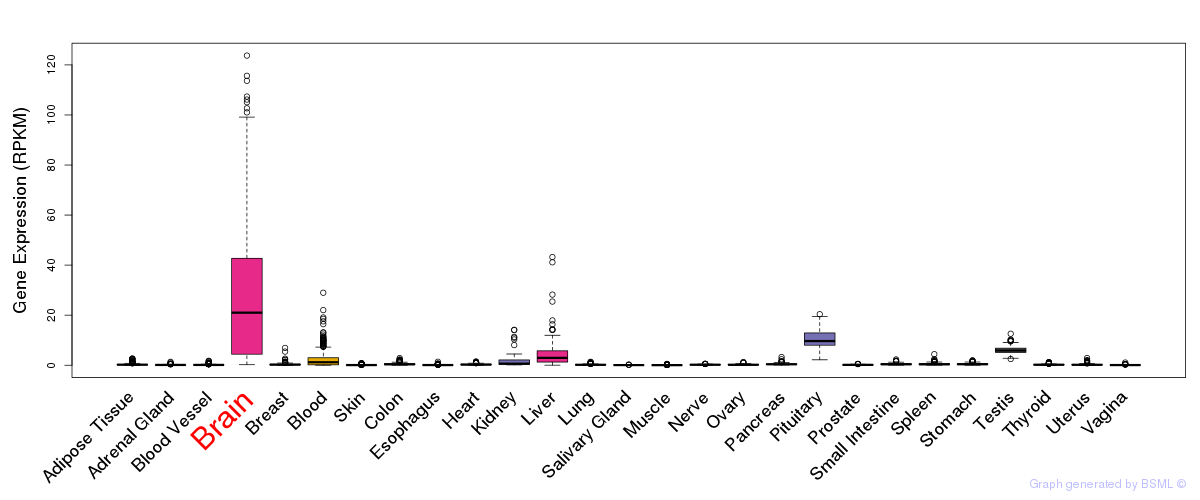
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CAPN3 | CANP3 | CANPL3 | LGMD2 | LGMD2A | MGC10767 | MGC11121 | MGC14344 | MGC4403 | nCL-1 | p94 | calpain 3, (p94) | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| CCNK | CPR4 | MGC9113 | cyclin K | Two-hybrid | BioGRID | 16189514 |
| CENPO | CENP-O | MGC11266 | centromere protein O | Two-hybrid | BioGRID | 16189514 |
| DFFA | DFF-45 | DFF1 | ICAD | DNA fragmentation factor, 45kDa, alpha polypeptide | Two-hybrid | BioGRID | 16189514 |
| DGCR6L | FLJ10666 | DiGeorge syndrome critical region gene 6-like | Two-hybrid | BioGRID | 16189514 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| GTPBP10 | DKFZp686A10121 | FLJ38242 | MGC104191 | ObgH2 | UG0751c10 | GTP-binding protein 10 (putative) | Two-hybrid | BioGRID | 16189514 |
| KIAA1267 | DKFZp686P06109 | DKFZp727C091 | MGC102843 | KIAA1267 | Two-hybrid | BioGRID | 16189514 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| NEK6 | SID6-1512 | NIMA (never in mitosis gene a)-related kinase 6 | Two-hybrid | BioGRID | 16189514 |
| NOC4L | MGC3162 | NOC4 | nucleolar complex associated 4 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| PSMA1 | HC2 | MGC14542 | MGC14575 | MGC14751 | MGC1667 | MGC21459 | MGC22853 | MGC23915 | NU | PROS30 | proteasome (prosome, macropain) subunit, alpha type, 1 | Two-hybrid | BioGRID | 16189514 |
| RCOR3 | FLJ10876 | FLJ16298 | RP11-318L16.1 | REST corepressor 3 | Two-hybrid | BioGRID | 16189514 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | Two-hybrid | BioGRID | 16189514 |
| TEX11 | TGC1 | TSGA3 | testis expressed 11 | Two-hybrid | BioGRID | 16189514 |
| VTA1 | C6orf55 | DRG-1 | DRG1 | FLJ27228 | HSPC228 | LIP5 | My012 | SBP1 | Vps20-associated 1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| WDYHV1 | C8orf32 | FLJ10204 | WDYHV motif containing 1 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |