Gene Page: UGT1A4
Summary ?
| GeneID | 54657 |
| Symbol | UGT1A4 |
| Synonyms | HUG-BR2|UDPGT|UDPGT 1-4|UGT-1D|UGT1-04|UGT1.4|UGT1A4S|UGT1D |
| Description | UDP glucuronosyltransferase family 1 member A4 |
| Reference | MIM:606429|HGNC:HGNC:12536|Ensembl:ENSG00000244474|HPRD:08402|Vega:OTTHUMG00000059119 |
| Gene type | protein-coding |
| Map location | 2q37 |
| Pascal p-value | 0.762 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 0 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|
Section II. Transcriptome annotation
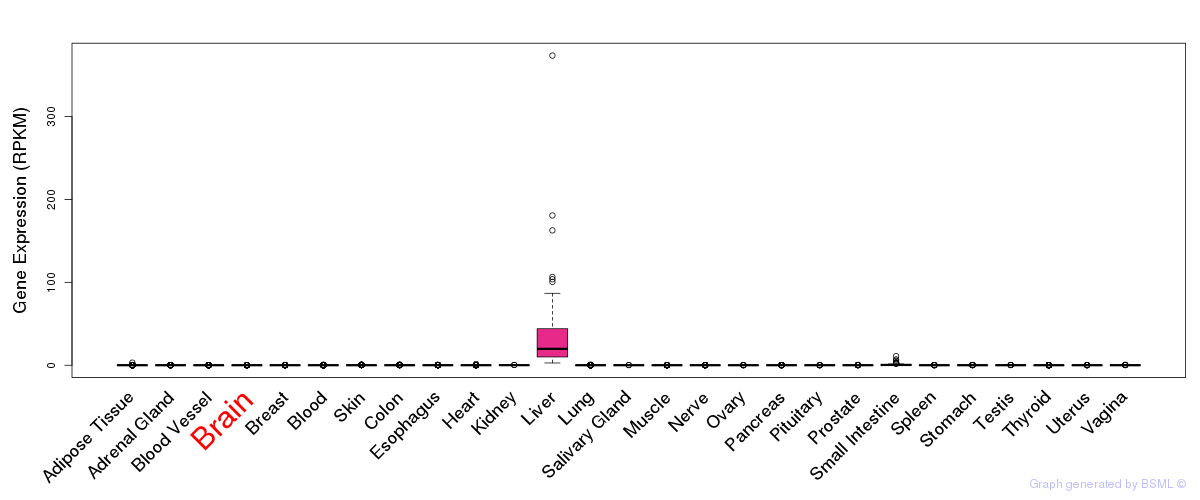
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016758 | transferase activity, transferring hexosyl groups | IEA | - | |
| GO:0015020 | glucuronosyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008152 | metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005792 | microsome | IEA | - | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005783 | endoplasmic reticulum | TAS | 1898728 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PENTOSE AND GLUCURONATE INTERCONVERSIONS | 28 | 17 | All SZGR 2.0 genes in this pathway |
| KEGG ASCORBATE AND ALDARATE METABOLISM | 25 | 17 | All SZGR 2.0 genes in this pathway |
| KEGG STEROID HORMONE BIOSYNTHESIS | 55 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG STARCH AND SUCROSE METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG RETINOL METABOLISM | 64 | 37 | All SZGR 2.0 genes in this pathway |
| KEGG PORPHYRIN AND CHLOROPHYLL METABOLISM | 41 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG METABOLISM OF XENOBIOTICS BY CYTOCHROME P450 | 70 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM CYTOCHROME P450 | 72 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG DRUG METABOLISM OTHER ENZYMES | 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCURONIDATION | 18 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE II CONJUGATION | 70 | 42 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |