Gene Page: RNF43
Summary ?
| GeneID | 54894 |
| Symbol | RNF43 |
| Synonyms | RNF124|URCC |
| Description | ring finger protein 43 |
| Reference | MIM:612482|HGNC:HGNC:18505|Ensembl:ENSG00000108375|HPRD:07896|Vega:OTTHUMG00000179077 |
| Gene type | protein-coding |
| Map location | 17q22 |
| Pascal p-value | 0.006 |
| Sherlock p-value | 0.046 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | RNF43 | 54894 | 3.556E-5 | trans | ||
| rs16841750 | chr2 | 158288461 | RNF43 | 54894 | 0.16 | trans | ||
| rs7584986 | chr2 | 184111432 | RNF43 | 54894 | 0.11 | trans | ||
| rs7306102 | chr12 | 24607037 | RNF43 | 54894 | 0.11 | trans | ||
| rs16955618 | chr15 | 29937543 | RNF43 | 54894 | 2.316E-8 | trans |
Section II. Transcriptome annotation
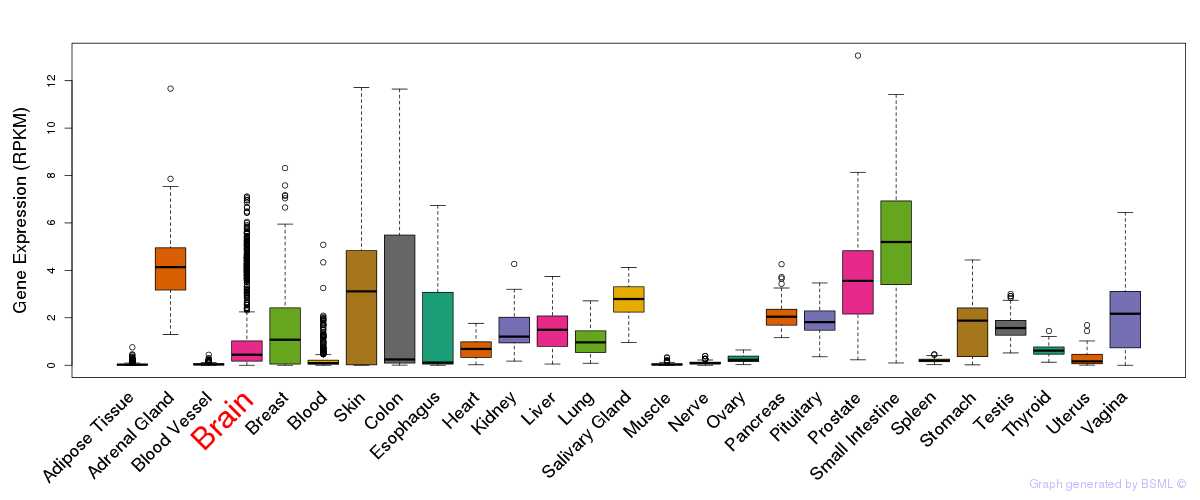
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KITLG | 0.88 | 0.47 |
| LHX9 | 0.86 | 0.25 |
| EPHA1 | 0.85 | 0.31 |
| SLITRK6 | 0.84 | 0.48 |
| GBX2 | 0.83 | 0.32 |
| ANO2 | 0.82 | 0.52 |
| FOXP2 | 0.81 | 0.37 |
| KRT18P19 | 0.81 | 0.19 |
| FIGN | 0.80 | 0.47 |
| PDE5A | 0.80 | 0.43 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CIDEA | -0.20 | -0.15 |
| KIAA1614 | -0.20 | -0.04 |
| S100A1 | -0.19 | -0.08 |
| AF347015.33 | -0.19 | -0.18 |
| AC018755.7 | -0.19 | -0.10 |
| EMX1 | -0.19 | -0.12 |
| PC | -0.19 | -0.09 |
| AF347015.2 | -0.19 | -0.17 |
| AIF1L | -0.18 | -0.10 |
| MT-CO2 | -0.18 | -0.14 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP | 121 | 45 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| IVANOV MUTATED IN COLON CANCER | 13 | 9 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C DN | 59 | 39 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G6 UP | 65 | 43 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 5 | 18 | 11 | All SZGR 2.0 genes in this pathway |