Gene Page: WHSC1L1
Summary ?
| GeneID | 54904 |
| Symbol | WHSC1L1 |
| Synonyms | NSD3|WHISTLE|pp14328 |
| Description | Wolf-Hirschhorn syndrome candidate 1-like 1 |
| Reference | MIM:607083|HGNC:HGNC:12767|Ensembl:ENSG00000147548|HPRD:06155|Vega:OTTHUMG00000165115 |
| Gene type | protein-coding |
| Map location | 8p11.2 |
| Pascal p-value | 1.364E-6 |
| Sherlock p-value | 0.478 |
| Fetal beta | 1.558 |
| DMG | 1 (# studies) |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01609705 | 8 | 38240669 | WHSC1L1 | 4.25E-9 | -0.006 | 2.55E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
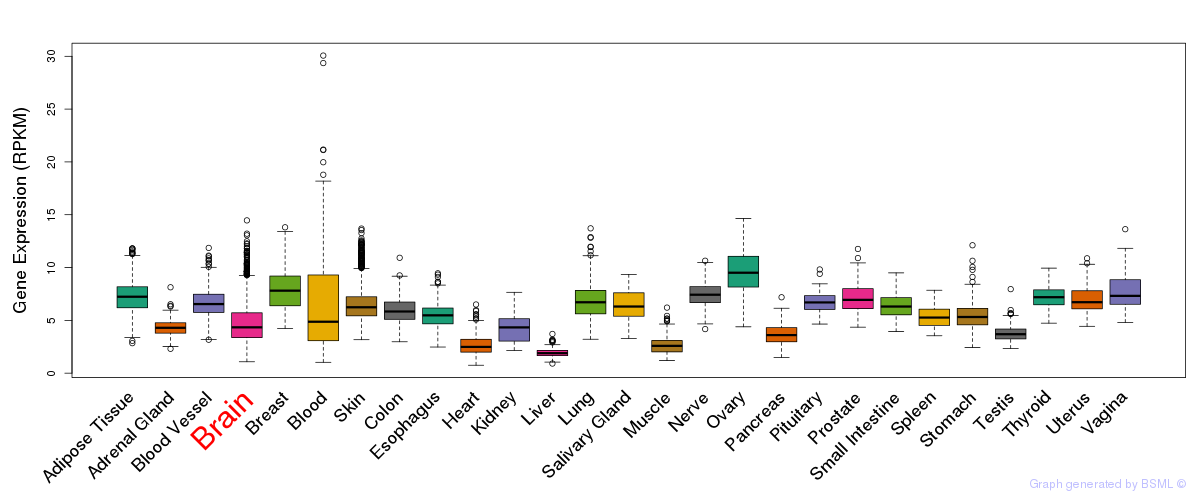
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C11orf59 | 0.90 | 0.82 |
| COX8A | 0.90 | 0.81 |
| MPG | 0.89 | 0.76 |
| C17orf90 | 0.89 | 0.77 |
| ZNF593 | 0.87 | 0.70 |
| MRP63 | 0.86 | 0.76 |
| NDUFA7 | 0.86 | 0.63 |
| SIVA1 | 0.85 | 0.76 |
| ISOC2 | 0.85 | 0.74 |
| MRPL55 | 0.85 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.39 | -0.46 |
| RBM25 | -0.38 | -0.35 |
| ZNHIT6 | -0.37 | -0.28 |
| AC010300.1 | -0.37 | -0.50 |
| ZNF326 | -0.36 | -0.31 |
| CCDC55 | -0.36 | -0.28 |
| AC005921.3 | -0.34 | -0.47 |
| Z83840.4 | -0.34 | -0.32 |
| AF347015.18 | -0.34 | -0.30 |
| SFRS12 | -0.33 | -0.27 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSINE DEGRADATION | 44 | 29 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 9 | 8 | 6 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |