Gene Page: PIGX
Summary ?
| GeneID | 54965 |
| Symbol | PIGX |
| Synonyms | PIG-X |
| Description | phosphatidylinositol glycan anchor biosynthesis class X |
| Reference | MIM:610276|HGNC:HGNC:26046|Ensembl:ENSG00000163964|HPRD:07921|Vega:OTTHUMG00000155535 |
| Gene type | protein-coding |
| Map location | 3q29 |
| Pascal p-value | 0.314 |
| Sherlock p-value | 0.229 |
| Fetal beta | -0.404 |
| eGene | Anterior cingulate cortex BA24 Cortex Hypothalamus Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2716734 | chr2 | 39947720 | PIGX | 54965 | 0.2 | trans | ||
| rs2716736 | chr2 | 39947946 | PIGX | 54965 | 0.2 | trans | ||
| rs12255258 | chr10 | 106306927 | PIGX | 54965 | 0.07 | trans | ||
| rs76114543 | 3 | 196513710 | PIGX | ENSG00000163964.9 | 7.182E-8 | 0.01 | 74481 | gtex_brain_ba24 |
| rs34087742 | 3 | 196514583 | PIGX | ENSG00000163964.9 | 9.057E-7 | 0.01 | 75354 | gtex_brain_ba24 |
| rs34640252 | 3 | 196514873 | PIGX | ENSG00000163964.9 | 9.057E-7 | 0.01 | 75644 | gtex_brain_ba24 |
| rs11923075 | 3 | 196515332 | PIGX | ENSG00000163964.9 | 3.354E-7 | 0.01 | 76103 | gtex_brain_ba24 |
| rs4916548 | 3 | 196516661 | PIGX | ENSG00000163964.9 | 9.057E-7 | 0.01 | 77432 | gtex_brain_ba24 |
| rs4916549 | 3 | 196516663 | PIGX | ENSG00000163964.9 | 3.354E-7 | 0.01 | 77434 | gtex_brain_ba24 |
| rs6796039 | 3 | 196517754 | PIGX | ENSG00000163964.9 | 9.057E-7 | 0.01 | 78525 | gtex_brain_ba24 |
| rs12497912 | 3 | 196518026 | PIGX | ENSG00000163964.9 | 9.057E-7 | 0.01 | 78797 | gtex_brain_ba24 |
| rs12494636 | 3 | 196518079 | PIGX | ENSG00000163964.9 | 8.571E-7 | 0.01 | 78850 | gtex_brain_ba24 |
| rs13062353 | 3 | 196518611 | PIGX | ENSG00000163964.9 | 5.109E-7 | 0.01 | 79382 | gtex_brain_ba24 |
| rs13082070 | 3 | 196518791 | PIGX | ENSG00000163964.9 | 2.423E-7 | 0.01 | 79562 | gtex_brain_ba24 |
| rs13062709 | 3 | 196518852 | PIGX | ENSG00000163964.9 | 4.182E-7 | 0.01 | 79623 | gtex_brain_ba24 |
| rs35314545 | 3 | 196443947 | PIGX | ENSG00000163964.9 | 1.172E-6 | 0.03 | 4718 | gtex_brain_putamen_basal |
| rs71323743 | 3 | 196445193 | PIGX | ENSG00000163964.9 | 1.018E-6 | 0.03 | 5964 | gtex_brain_putamen_basal |
| rs1568603 | 3 | 196453467 | PIGX | ENSG00000163964.9 | 1.556E-6 | 0.03 | 14238 | gtex_brain_putamen_basal |
| rs71323744 | 3 | 196455682 | PIGX | ENSG00000163964.9 | 1.203E-6 | 0.03 | 16453 | gtex_brain_putamen_basal |
| rs56936358 | 3 | 196459988 | PIGX | ENSG00000163964.9 | 1.203E-6 | 0.03 | 20759 | gtex_brain_putamen_basal |
| rs34830136 | 3 | 196460680 | PIGX | ENSG00000163964.9 | 1.203E-6 | 0.03 | 21451 | gtex_brain_putamen_basal |
| rs4916542 | 3 | 196461390 | PIGX | ENSG00000163964.9 | 1.203E-6 | 0.03 | 22161 | gtex_brain_putamen_basal |
| rs4916543 | 3 | 196462949 | PIGX | ENSG00000163964.9 | 1.203E-6 | 0.03 | 23720 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
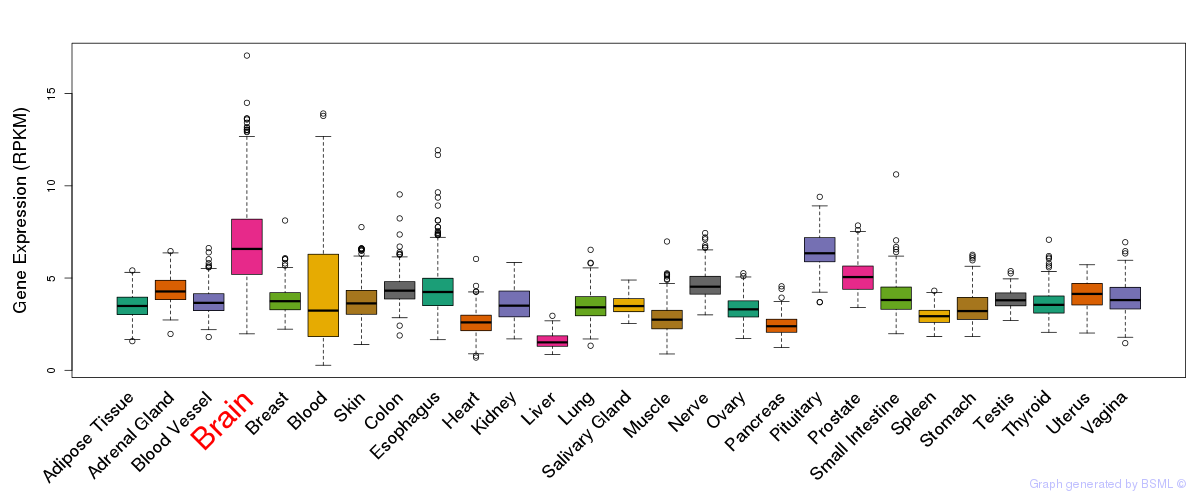
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSYLPHOSPHATIDYLINOSITOL GPI ANCHOR BIOSYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | 17 | 12 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |