Gene Page: AUP1
Summary ?
| GeneID | 550 |
| Symbol | AUP1 |
| Synonyms | - |
| Description | ancient ubiquitous protein 1 |
| Reference | MIM:602434|HGNC:HGNC:891|Ensembl:ENSG00000115307|HPRD:03892|Vega:OTTHUMG00000129964 |
| Gene type | protein-coding |
| Map location | 2p13 |
| Pascal p-value | 0.205 |
| Sherlock p-value | 0.521 |
| Fetal beta | 0.384 |
| DMG | 1 (# studies) |
| eGene | Hypothalamus |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06721411 | 2 | 74753759 | AUP1 | 2.67E-6 | 0.009 | 0.023 | DMG:Montano_2016 |
Section II. Transcriptome annotation
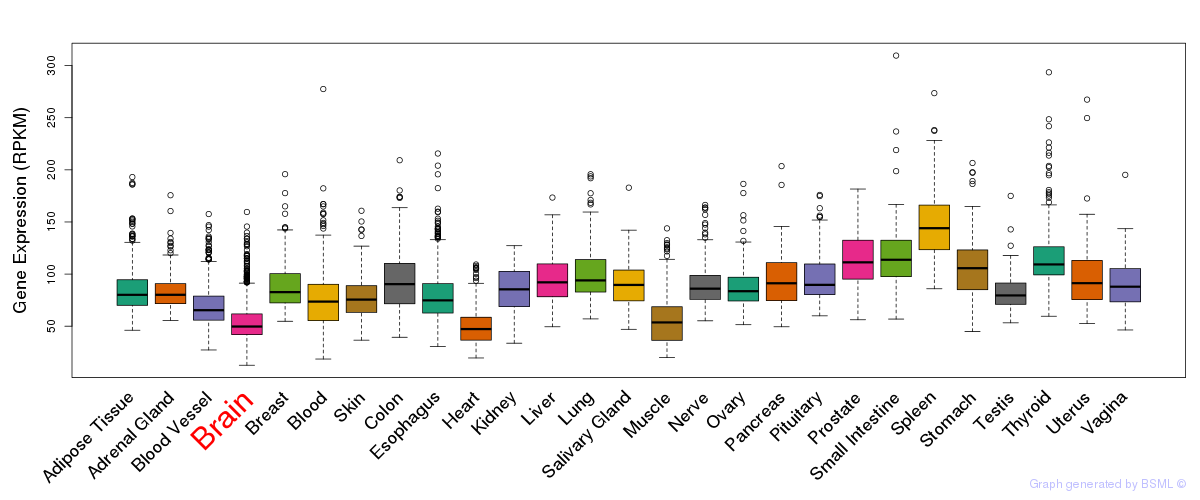
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HOMER2 | 0.65 | 0.47 |
| PITPNM1 | 0.62 | 0.49 |
| GALNT14 | 0.60 | 0.41 |
| ZDHHC22 | 0.59 | 0.41 |
| PAPPA | 0.59 | 0.20 |
| PTPN3 | 0.59 | 0.31 |
| NOD2 | 0.58 | 0.10 |
| CHRNA4 | 0.58 | 0.51 |
| TPH2 | 0.57 | 0.14 |
| NTNG1 | 0.57 | 0.32 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OAF | -0.26 | -0.36 |
| GRIN2C | -0.26 | -0.34 |
| AIF1L | -0.26 | -0.37 |
| HEPN1 | -0.25 | -0.36 |
| AF347015.2 | -0.25 | -0.36 |
| NDUFA4L2 | -0.25 | -0.33 |
| S100A1 | -0.25 | -0.35 |
| SELENBP1 | -0.25 | -0.34 |
| WIF1 | -0.24 | -0.32 |
| FXYD1 | -0.24 | -0.35 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATG12 | APG12 | APG12L | FBR93 | HAPG12 | ATG12 autophagy related 12 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Affinity Capture-MS | BioGRID | 17353931 |
| ITGA1 | CD49a | VLA1 | integrin, alpha 1 | - | HPRD,BioGRID | 12042322 |
| ITGA2 | BR | CD49B | GPIa | VLA-2 | VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | - | HPRD,BioGRID | 12042322 |
| ITGA2B | CD41 | CD41B | GP2B | GPIIb | GTA | HPA3 | integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | - | HPRD,BioGRID | 12042322 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | - | HPRD,BioGRID | 12042322 |
| ITGAM | CD11B | CR3A | MAC-1 | MAC1A | MGC117044 | MO1A | SLEB6 | integrin, alpha M (complement component 3 receptor 3 subunit) | - | HPRD,BioGRID | 12042322 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | - | HPRD,BioGRID | 12042322 |
| KPTN | 2E4 | kaptin (actin binding protein) | Affinity Capture-MS | BioGRID | 17353931 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | Affinity Capture-MS | BioGRID | 17353931 |
| UBE2G2 | UBC7 | ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |