Gene Page: PID1
Summary ?
| GeneID | 55022 |
| Symbol | PID1 |
| Synonyms | HMFN2073|NYGGF4|P-CLI1|PCLI1 |
| Description | phosphotyrosine interaction domain containing 1 |
| Reference | MIM:612930|HGNC:HGNC:26084|Ensembl:ENSG00000153823|HPRD:07944|Vega:OTTHUMG00000133191 |
| Gene type | protein-coding |
| Map location | 2q36.3 |
| Pascal p-value | 0.012 |
| Sherlock p-value | 0.208 |
| Fetal beta | 0.424 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2809658 | chr1 | 43050395 | PID1 | 55022 | 0.03 | trans |
Section II. Transcriptome annotation
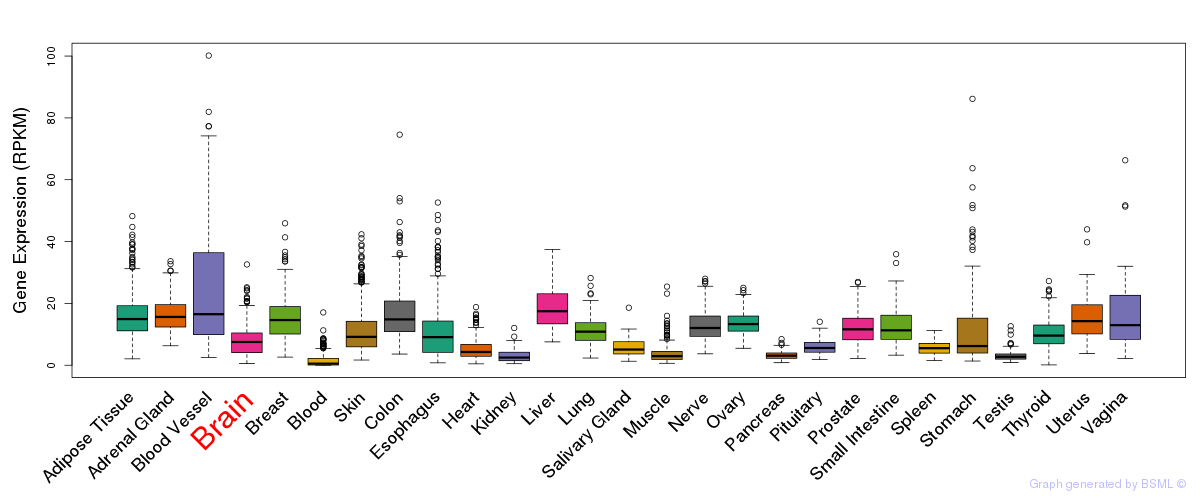
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPRJ | 0.90 | 0.91 |
| PHLPP2 | 0.87 | 0.87 |
| ENTPD4 | 0.87 | 0.88 |
| PNMAL1 | 0.87 | 0.87 |
| AP3M2 | 0.87 | 0.88 |
| ABR | 0.87 | 0.87 |
| TGOLN2 | 0.86 | 0.87 |
| SCN3B | 0.86 | 0.85 |
| HTT | 0.85 | 0.87 |
| MEF2A | 0.85 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.53 | -0.47 |
| AL139819.3 | -0.50 | -0.51 |
| AF347015.18 | -0.49 | -0.45 |
| HIGD1B | -0.49 | -0.45 |
| AP002478.3 | -0.48 | -0.47 |
| MT-CO2 | -0.48 | -0.45 |
| AF347015.8 | -0.48 | -0.44 |
| AC098691.2 | -0.47 | -0.48 |
| GNG11 | -0.47 | -0.47 |
| C1orf54 | -0.47 | -0.51 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FOURNIER ACINAR DEVELOPMENT EARLY UP | 21 | 19 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX5 UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |