Gene Page: THAP1
Summary ?
| GeneID | 55145 |
| Symbol | THAP1 |
| Synonyms | DYT6 |
| Description | THAP domain containing, apoptosis associated protein 1 |
| Reference | MIM:609520|HGNC:HGNC:20856|Ensembl:ENSG00000131931|HPRD:15496|Vega:OTTHUMG00000165276 |
| Gene type | protein-coding |
| Map location | 8p11.21 |
| Pascal p-value | 0.029 |
| Fetal beta | 1.153 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03301282 | 8 | 42698936 | THAP1 | 2.11E-5 | -0.588 | 0.017 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
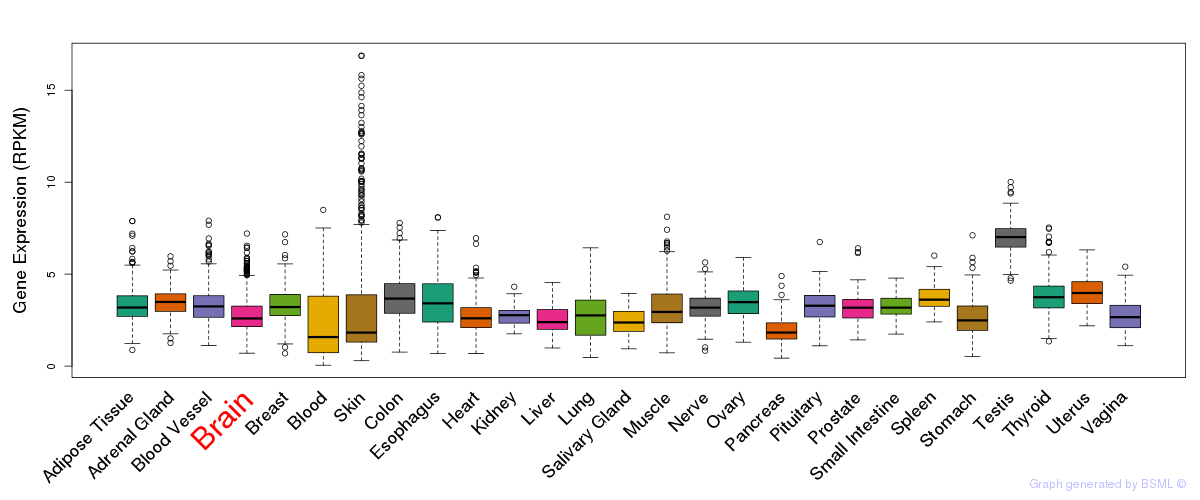
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP2B1 | ADTB2 | AP105B | AP2-BETA | CLAPB1 | DKFZp781K0743 | adaptor-related protein complex 2, beta 1 subunit | Two-hybrid | BioGRID | 16189514 |
| C1orf35 | MGC4174 | MMTAG2 | chromosome 1 open reading frame 35 | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| GRINL1A | DKFZp586F1918 | glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A | Two-hybrid | BioGRID | 16189514 |
| NKAP | FLJ22626 | NFKB activating protein | Two-hybrid | BioGRID | 16189514 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | Two-hybrid | BioGRID | 16189514 |
| PAWR | PAR4 | Par-4 | PRKC, apoptosis, WT1, regulator | - | HPRD,BioGRID | 12717420 |
| PHF1 | MTF2L2 | PCL1 | PHF2 | PHD finger protein 1 | Two-hybrid | BioGRID | 16189514 |
| RALYL | HNRPCL3 | RALY RNA binding protein-like | Two-hybrid | BioGRID | 16189514 |
| STRBP | DKFZp434N214 | FLJ11307 | FLJ14223 | FLJ14984 | ILF3L | MGC21529 | MGC3405 | SPNR | p74 | spermatid perinuclear RNA binding protein | Two-hybrid | BioGRID | 16189514 |
| THAP1 | FLJ10477 | MGC33014 | THAP domain containing, apoptosis associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | Two-hybrid | BioGRID | 16189514 |
| ZCCHC10 | FLJ20094 | zinc finger, CCHC domain containing 10 | Two-hybrid | BioGRID | 16189514 |
| ZNF408 | FLJ12827 | zinc finger protein 408 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |