Gene Page: PPP6C
Summary ?
| GeneID | 5537 |
| Symbol | PPP6C |
| Synonyms | PP6|PP6C |
| Description | protein phosphatase 6 catalytic subunit |
| Reference | MIM:612725|HGNC:HGNC:9323|Ensembl:ENSG00000119414|HPRD:02141|Vega:OTTHUMG00000020671 |
| Gene type | protein-coding |
| Map location | 9q33.3 |
| Pascal p-value | 0.318 |
| Sherlock p-value | 0.587 |
| Fetal beta | -0.143 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.866 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03801144 | 9 | 127951957 | PPP6C | -0.018 | 0.83 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs620677 | 9 | 127819261 | PPP6C | ENSG00000119414.7 | 4.09463E-6 | 0.01 | 132957 | gtex_brain_putamen_basal |
| rs687654 | 9 | 127833905 | PPP6C | ENSG00000119414.7 | 1.90289E-6 | 0.01 | 118313 | gtex_brain_putamen_basal |
| rs650599 | 9 | 127849318 | PPP6C | ENSG00000119414.7 | 3.38774E-7 | 0.01 | 102900 | gtex_brain_putamen_basal |
| rs58809143 | 9 | 127961613 | PPP6C | ENSG00000119414.7 | 1.26497E-6 | 0.01 | -9395 | gtex_brain_putamen_basal |
| rs405631 | 9 | 127968496 | PPP6C | ENSG00000119414.7 | 2.1975E-6 | 0.01 | -16278 | gtex_brain_putamen_basal |
| rs500438 | 9 | 127970845 | PPP6C | ENSG00000119414.7 | 1.71951E-6 | 0.01 | -18627 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
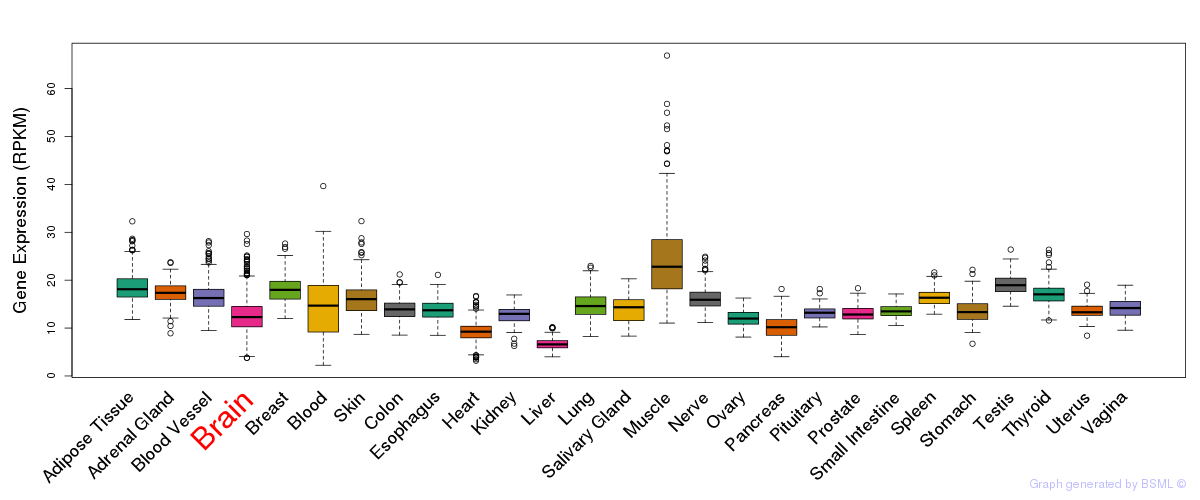
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM164A | 0.94 | 0.96 |
| RSG17 | 0.94 | 0.95 |
| TTC13 | 0.93 | 0.95 |
| HMGCR | 0.93 | 0.94 |
| RNF2 | 0.92 | 0.95 |
| PRKAR2B | 0.92 | 0.95 |
| ARPC5 | 0.91 | 0.95 |
| RUFY3 | 0.91 | 0.93 |
| SH3GLB1 | 0.91 | 0.90 |
| NCUBE1 | 0.91 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.72 | -0.81 |
| AIFM3 | -0.71 | -0.80 |
| TSC22D4 | -0.69 | -0.81 |
| MT-CO2 | -0.69 | -0.88 |
| AF347015.27 | -0.69 | -0.85 |
| HEPN1 | -0.69 | -0.78 |
| AF347015.31 | -0.68 | -0.86 |
| AF347015.33 | -0.68 | -0.86 |
| ALDOC | -0.67 | -0.72 |
| S100B | -0.67 | -0.81 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 16716191 | |
| GO:0004722 | protein serine/threonine phosphatase activity | IDA | 16716191 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | TAS | 9013334 | |
| GO:0006470 | protein amino acid dephosphorylation | IDA | 16716191 | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IDA | 16716191 | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 261 | 267 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA | ||||
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-15/16/195/424/497 | 95 | 101 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 89 | 95 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-208 | 245 | 251 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-214 | 97 | 103 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-221/222 | 267 | 273 | m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-31 | 113 | 119 | m8 | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-335 | 272 | 278 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-495 | 156 | 162 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-499 | 245 | 251 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 88 | 94 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.