Gene Page: MCM10
Summary ?
| GeneID | 55388 |
| Symbol | MCM10 |
| Synonyms | CNA43|DNA43|PRO2249 |
| Description | minichromosome maintenance 10 replication initiation factor |
| Reference | MIM:609357|HGNC:HGNC:18043|Ensembl:ENSG00000065328|HPRD:11297|Vega:OTTHUMG00000017694 |
| Gene type | protein-coding |
| Map location | 10p13 |
| Pascal p-value | 0.353 |
| Fetal beta | 4.35 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26884570 | 10 | 13204110 | MCM10 | 3.221E-4 | -0.471 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11124283 | chr2 | 32767095 | MCM10 | 55388 | 0.16 | trans |
Section II. Transcriptome annotation
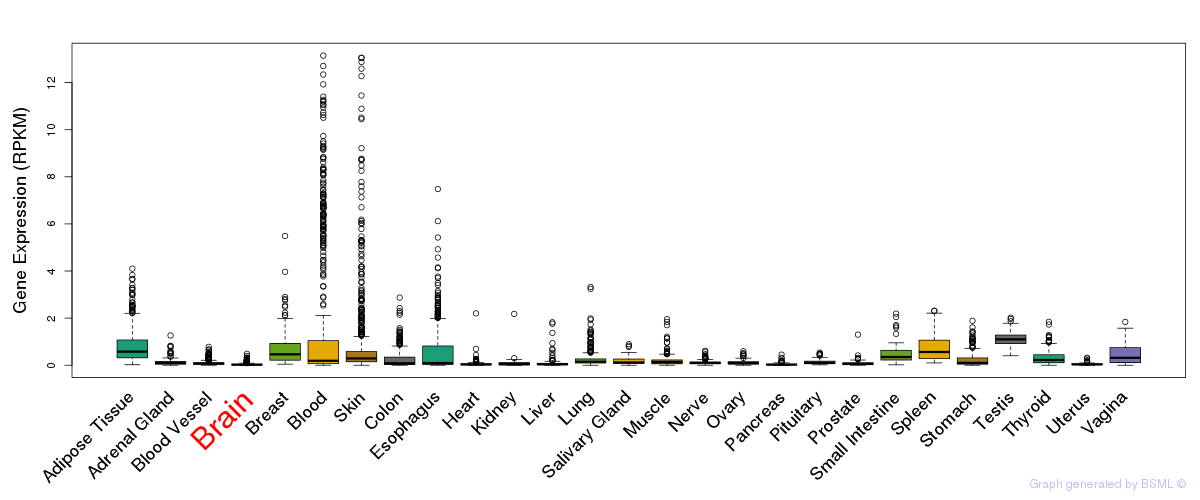
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| CCDC5 | FLJ21094 | FLJ40084 | HEI-C | HsT1461 | coiled-coil domain containing 5 (spindle associated) | - | HPRD | 15232106 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD | 15232106 |
| CCND3 | - | cyclin D3 | Affinity Capture-Western | BioGRID | 16189514 |
| CCND3 | - | cyclin D3 | - | HPRD | 15232106 |
| CDC45L | CDC45 | CDC45L2 | PORC-PI-1 | CDC45 cell division cycle 45-like (S. cerevisiae) | - | HPRD | 15232106 |
| CDC6 | CDC18L | HsCDC18 | HsCDC6 | cell division cycle 6 homolog (S. cerevisiae) | - | HPRD | 15232106 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD | 15232106 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | - | HPRD | 15232106 |
| CEP70 | BITE | FLJ13036 | centrosomal protein 70kDa | Two-hybrid | BioGRID | 16189514 |
| CEP72 | FLJ10565 | KIAA1519 | MGC5307 | centrosomal protein 72kDa | Two-hybrid | BioGRID | 16189514 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| HOOK2 | FLJ26218 | HK2 | hook homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| KRT15 | CK15 | K15 | K1CO | keratin 15 | Two-hybrid | BioGRID | 16189514 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD,BioGRID | 11095689 |15232106 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD | 15232106 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | - | HPRD,BioGRID | 11095689 |15232106 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | - | HPRD,BioGRID | 11095689 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | in vitro in vivo Two-hybrid | BioGRID | 11095689 |16189514 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | - | HPRD | 11095689 |15232106 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD,BioGRID | 11095689 |15232106 |
| ORC2L | ORC2 | origin recognition complex, subunit 2-like (yeast) | - | HPRD,BioGRID | 11095689 |
| ORC3L | LAT | LATHEO | ORC3 | origin recognition complex, subunit 3-like (yeast) | - | HPRD | 15232106 |
| ORC4L | ORC4 | ORC4P | origin recognition complex, subunit 4-like (yeast) | - | HPRD | 15232106 |
| RP4-691N24.1 | FLJ11792 | KIAA0980 | NLP | dJ691N24.1 | ninein-like | Two-hybrid | BioGRID | 16189514 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | Two-hybrid | BioGRID | 16189514 |
| ZBTB43 | ZBTB22B | ZNF-X | ZNF297B | zinc finger and BTB domain containing 43 | Two-hybrid | BioGRID | 16189514 |
| ZBTB8 | BOZF1 | FLJ90065 | MGC17919 | zinc finger and BTB domain containing 8 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | 38 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 2 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 DN | 65 | 39 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA DN | 74 | 45 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA PR UP | 45 | 20 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS RESPONSIVE TO ESTROGEN DN | 41 | 26 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D DN | 49 | 34 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| LEE EARLY T LYMPHOCYTE UP | 107 | 59 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS | 85 | 55 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |