Gene Page: PRELP
Summary ?
| GeneID | 5549 |
| Symbol | PRELP |
| Synonyms | MST161|MSTP161|SLRR2A |
| Description | proline/arginine-rich end leucine-rich repeat protein |
| Reference | MIM:601914|HGNC:HGNC:9357|Ensembl:ENSG00000188783|HPRD:03556|Vega:OTTHUMG00000035911 |
| Gene type | protein-coding |
| Map location | 1q32 |
| Sherlock p-value | 0.401 |
| Fetal beta | -0.767 |
| eGene | Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6668101 | chr1 | 36099199 | PRELP | 5549 | 0.04 | trans | ||
| rs1320385 | chr1 | 54064468 | PRELP | 5549 | 0.06 | trans | ||
| rs12036675 | chr1 | 54066317 | PRELP | 5549 | 0.06 | trans | ||
| rs17131472 | chr1 | 91962966 | PRELP | 5549 | 0.01 | trans | ||
| rs546975 | chr1 | 91963738 | PRELP | 5549 | 0.14 | trans | ||
| rs4953361 | chr2 | 46598567 | PRELP | 5549 | 0.04 | trans | ||
| rs1566993 | chr2 | 117105059 | PRELP | 5549 | 0.07 | trans | ||
| rs16825495 | chr2 | 134234401 | PRELP | 5549 | 0 | trans | ||
| rs7340510 | chr2 | 134267368 | PRELP | 5549 | 0.1 | trans | ||
| rs6730679 | chr2 | 134282345 | PRELP | 5549 | 0.1 | trans | ||
| rs2922180 | chr3 | 125280472 | PRELP | 5549 | 0 | trans | ||
| rs2976809 | chr3 | 125451738 | PRELP | 5549 | 0.04 | trans | ||
| rs2579326 | chr4 | 72131304 | PRELP | 5549 | 0.03 | trans | ||
| rs10050805 | chr5 | 30805352 | PRELP | 5549 | 0.1 | trans | ||
| rs317971 | chr5 | 66688724 | PRELP | 5549 | 0 | trans | ||
| rs248049 | chr5 | 126489320 | PRELP | 5549 | 0.01 | trans | ||
| rs17135501 | chr6 | 2704474 | PRELP | 5549 | 0.19 | trans | ||
| rs17135521 | chr6 | 2707631 | PRELP | 5549 | 0.19 | trans | ||
| snp_a-1787926 | 0 | PRELP | 5549 | 0.19 | trans | |||
| rs7747217 | chr6 | 3576943 | PRELP | 5549 | 0.07 | trans | ||
| rs9381511 | chr6 | 12931447 | PRELP | 5549 | 0 | trans | ||
| rs16889690 | chr6 | 34383787 | PRELP | 5549 | 0.18 | trans | ||
| rs16889851 | chr6 | 34401377 | PRELP | 5549 | 0.18 | trans | ||
| rs6934246 | chr6 | 35154494 | PRELP | 5549 | 6.311E-7 | trans | ||
| rs16871832 | chr6 | 44348256 | PRELP | 5549 | 0.05 | trans | ||
| rs4711782 | chr6 | 44363229 | PRELP | 5549 | 0.08 | trans | ||
| rs6455226 | chr6 | 67930295 | PRELP | 5549 | 0 | trans | ||
| rs12056282 | chr7 | 18882202 | PRELP | 5549 | 0.04 | trans | ||
| rs10085826 | chr7 | 42504690 | PRELP | 5549 | 0.05 | trans | ||
| rs6970701 | chr7 | 93087543 | PRELP | 5549 | 0.19 | trans | ||
| rs12111850 | chr7 | 93126467 | PRELP | 5549 | 0.19 | trans | ||
| rs17165529 | chr7 | 93144582 | PRELP | 5549 | 0.19 | trans | ||
| rs7841407 | chr8 | 9243427 | PRELP | 5549 | 0.15 | trans | ||
| rs680372 | chr8 | 36999626 | PRELP | 5549 | 0.01 | trans | ||
| rs7081666 | chr10 | 47673800 | PRELP | 5549 | 0.05 | trans | ||
| rs11006556 | chr10 | 61287609 | PRELP | 5549 | 0.01 | trans | ||
| rs12573318 | chr10 | 82909757 | PRELP | 5549 | 0.02 | trans | ||
| rs1159612 | chr10 | 85329057 | PRELP | 5549 | 4.348E-5 | trans | ||
| rs17653230 | chr10 | 120201621 | PRELP | 5549 | 0.14 | trans | ||
| rs10502229 | chr11 | 117979969 | PRELP | 5549 | 0.13 | trans | ||
| rs11049036 | chr12 | 27650292 | PRELP | 5549 | 0 | trans | ||
| rs7303819 | chr12 | 33990776 | PRELP | 5549 | 0 | trans | ||
| rs10879027 | chr12 | 70237156 | PRELP | 5549 | 0 | trans | ||
| rs2802244 | chr13 | 30081710 | PRELP | 5549 | 0.14 | trans | ||
| rs2802243 | chr13 | 30081748 | PRELP | 5549 | 0.14 | trans | ||
| rs1457420 | chr14 | 66958675 | PRELP | 5549 | 5.147E-4 | trans | ||
| rs1872761 | chr15 | 40003815 | PRELP | 5549 | 0.02 | trans | ||
| rs17137819 | chr16 | 5372534 | PRELP | 5549 | 0.01 | trans | ||
| rs16958302 | chr16 | 57647432 | PRELP | 5549 | 7.548E-6 | trans | ||
| snp_a-1955963 | 0 | PRELP | 5549 | 0.06 | trans | |||
| rs6034314 | chr20 | 15852130 | PRELP | 5549 | 0.14 | trans | ||
| rs4814410 | chr20 | 15867428 | PRELP | 5549 | 0.06 | trans | ||
| rs6095741 | chr20 | 48666589 | PRELP | 5549 | 5.302E-4 | trans | ||
| rs7265852 | chr20 | 48823717 | PRELP | 5549 | 0 | trans | ||
| rs5915713 | chrX | 4077883 | PRELP | 5549 | 2.306E-6 | trans | ||
| rs5944334 | chrX | 26177987 | PRELP | 5549 | 7.087E-4 | trans | ||
| rs5944341 | chrX | 26190706 | PRELP | 5549 | 7.087E-4 | trans | ||
| rs5991662 | chrX | 43335796 | PRELP | 5549 | 0.01 | trans | ||
| rs1545676 | chrX | 93530472 | PRELP | 5549 | 2.534E-9 | trans | ||
| rs1540914 | chrX | 99840177 | PRELP | 5549 | 0.07 | trans | ||
| rs5945924 | chrX | 102241590 | PRELP | 5549 | 0.2 | trans | ||
| rs112904762 | 1 | 204003496 | PRELP | ENSG00000188783.5 | 2.409E-6 | 0.03 | 558540 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
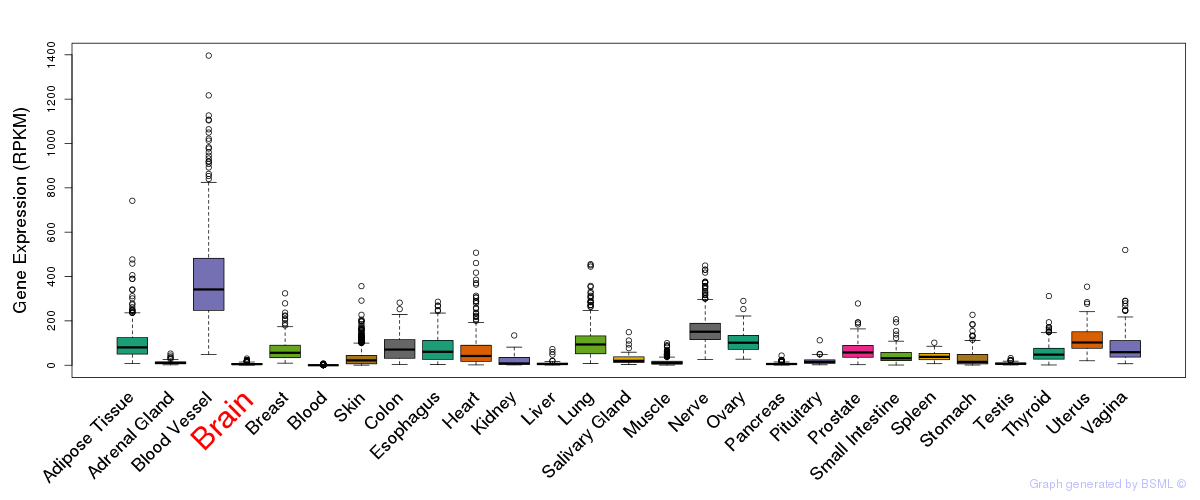
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DOCK3 | 0.93 | 0.94 |
| KIAA1467 | 0.91 | 0.93 |
| MADD | 0.91 | 0.92 |
| STXBP5 | 0.91 | 0.94 |
| DLGAP1 | 0.91 | 0.93 |
| PNMA2 | 0.90 | 0.91 |
| RAPGEF2 | 0.90 | 0.91 |
| HTT | 0.90 | 0.90 |
| BTBD9 | 0.90 | 0.90 |
| WDR7 | 0.89 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.60 | -0.66 |
| AP002478.3 | -0.58 | -0.65 |
| ACSF2 | -0.58 | -0.61 |
| FXYD1 | -0.57 | -0.56 |
| DBI | -0.57 | -0.64 |
| C1orf54 | -0.57 | -0.67 |
| HIGD1B | -0.57 | -0.59 |
| TLCD1 | -0.56 | -0.56 |
| C11orf67 | -0.56 | -0.60 |
| SAT1 | -0.56 | -0.64 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME KERATAN SULFATE BIOSYNTHESIS | 26 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE KERATIN METABOLISM | 30 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME KERATAN SULFATE DEGRADATION | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP | 50 | 27 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY DN | 12 | 8 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |