Gene Page: CHD7
Summary ?
| GeneID | 55636 |
| Symbol | CHD7 |
| Synonyms | CRG|HH5|IS3|KAL5 |
| Description | chromodomain helicase DNA binding protein 7 |
| Reference | MIM:608892|HGNC:HGNC:20626|Ensembl:ENSG00000171316|HPRD:10595|Vega:OTTHUMG00000165332 |
| Gene type | protein-coding |
| Map location | 8q12.2 |
| Pascal p-value | 0.032 |
| Fetal beta | 1.83 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13447530 | chr1 | 91983798 | CHD7 | 55636 | 0.19 | trans | ||
| rs7520276 | chr1 | 205659490 | CHD7 | 55636 | 0.11 | trans | ||
| rs16856160 | chr1 | 205661366 | CHD7 | 55636 | 0.11 | trans | ||
| rs17022378 | chr1 | 206654057 | CHD7 | 55636 | 0.06 | trans | ||
| rs17572651 | chr1 | 218943612 | CHD7 | 55636 | 0 | trans | ||
| rs6734526 | chr2 | 5616370 | CHD7 | 55636 | 0.11 | trans | ||
| rs6753473 | chr2 | 26526418 | CHD7 | 55636 | 0.01 | trans | ||
| rs2197597 | chr2 | 129638077 | CHD7 | 55636 | 0.15 | trans | ||
| rs2897468 | chr2 | 129711721 | CHD7 | 55636 | 0.14 | trans | ||
| rs16829545 | chr2 | 151977407 | CHD7 | 55636 | 1.914E-34 | trans | ||
| rs16841750 | chr2 | 158288461 | CHD7 | 55636 | 0.04 | trans | ||
| rs7584986 | chr2 | 184111432 | CHD7 | 55636 | 5.562E-5 | trans | ||
| rs6741060 | chr2 | 217169088 | CHD7 | 55636 | 9.897E-5 | trans | ||
| rs9810143 | chr3 | 5060209 | CHD7 | 55636 | 0 | trans | ||
| rs6797307 | chr3 | 8601563 | CHD7 | 55636 | 0 | trans | ||
| rs6773248 | chr3 | 174790333 | CHD7 | 55636 | 0.01 | trans | ||
| rs6797728 | chr3 | 174790417 | CHD7 | 55636 | 0.01 | trans | ||
| rs16882010 | chr4 | 11453741 | CHD7 | 55636 | 0.16 | trans | ||
| rs17415971 | chr4 | 14272434 | CHD7 | 55636 | 0.12 | trans | ||
| snp_a-1815771 | 0 | CHD7 | 55636 | 0.02 | trans | |||
| rs17020035 | chr4 | 93873810 | CHD7 | 55636 | 0.01 | trans | ||
| rs17020058 | chr4 | 93915972 | CHD7 | 55636 | 0.02 | trans | ||
| rs17020066 | chr4 | 93921055 | CHD7 | 55636 | 0.02 | trans | ||
| rs17008119 | chr4 | 125321425 | CHD7 | 55636 | 0.13 | trans | ||
| rs7692715 | chr4 | 125335209 | CHD7 | 55636 | 0.02 | trans | ||
| rs1480540 | chr4 | 171103335 | CHD7 | 55636 | 0.15 | trans | ||
| rs1396222 | chr4 | 173279496 | CHD7 | 55636 | 0.15 | trans | ||
| rs4488887 | chr4 | 177695833 | CHD7 | 55636 | 0.12 | trans | ||
| rs10474151 | chr5 | 84286402 | CHD7 | 55636 | 0.03 | trans | ||
| rs13360496 | 0 | CHD7 | 55636 | 0 | trans | |||
| rs13191953 | chr6 | 24151042 | CHD7 | 55636 | 0.03 | trans | ||
| rs12196880 | chr6 | 24313746 | CHD7 | 55636 | 0.08 | trans | ||
| rs16890367 | chr6 | 38078448 | CHD7 | 55636 | 0.04 | trans | ||
| rs17149888 | chr7 | 24738371 | CHD7 | 55636 | 0.15 | trans | ||
| rs10253181 | chr7 | 26563022 | CHD7 | 55636 | 0.11 | trans | ||
| rs12706918 | chr7 | 80266147 | CHD7 | 55636 | 0 | trans | ||
| rs4841493 | chr8 | 6521431 | CHD7 | 55636 | 0.1 | trans | ||
| rs6996695 | chr8 | 77540580 | CHD7 | 55636 | 0.07 | trans | ||
| rs11139334 | chr9 | 84209393 | CHD7 | 55636 | 9.232E-4 | trans | ||
| rs2393316 | chr10 | 59333070 | CHD7 | 55636 | 5.988E-4 | trans | ||
| rs17124813 | chr10 | 110347464 | CHD7 | 55636 | 0.03 | trans | ||
| rs11061703 | chr12 | 1436977 | CHD7 | 55636 | 0.04 | trans | ||
| rs9669370 | chr12 | 1466349 | CHD7 | 55636 | 0.01 | trans | ||
| rs7337967 | chr13 | 53519044 | CHD7 | 55636 | 0.01 | trans | ||
| rs4112777 | chr13 | 99281457 | CHD7 | 55636 | 0 | trans | ||
| rs9989228 | chr14 | 37809252 | CHD7 | 55636 | 0 | trans | ||
| rs16955618 | chr15 | 29937543 | CHD7 | 55636 | 5.44E-29 | trans | ||
| rs2077735 | chr15 | 58479024 | CHD7 | 55636 | 0 | trans | ||
| rs4795850 | chr17 | 32383815 | CHD7 | 55636 | 0.07 | trans | ||
| rs4795851 | chr17 | 32384050 | CHD7 | 55636 | 0.06 | trans | ||
| rs1003837 | chr17 | 32453154 | CHD7 | 55636 | 0.19 | trans | ||
| rs11873184 | chr18 | 1584081 | CHD7 | 55636 | 9.684E-4 | trans | ||
| rs12458173 | chr18 | 31430166 | CHD7 | 55636 | 0.05 | trans | ||
| rs17782355 | chr19 | 55807585 | CHD7 | 55636 | 0.12 | trans | ||
| rs7274477 | chr20 | 1676942 | CHD7 | 55636 | 0.05 | trans | ||
| rs6046679 | chr20 | 20146874 | CHD7 | 55636 | 0.16 | trans | ||
| rs4813543 | chr20 | 25004070 | CHD7 | 55636 | 0.07 | trans | ||
| rs1041786 | chr21 | 22617710 | CHD7 | 55636 | 0 | trans | ||
| rs3829726 | chrX | 72666188 | CHD7 | 55636 | 0.05 | trans |
Section II. Transcriptome annotation
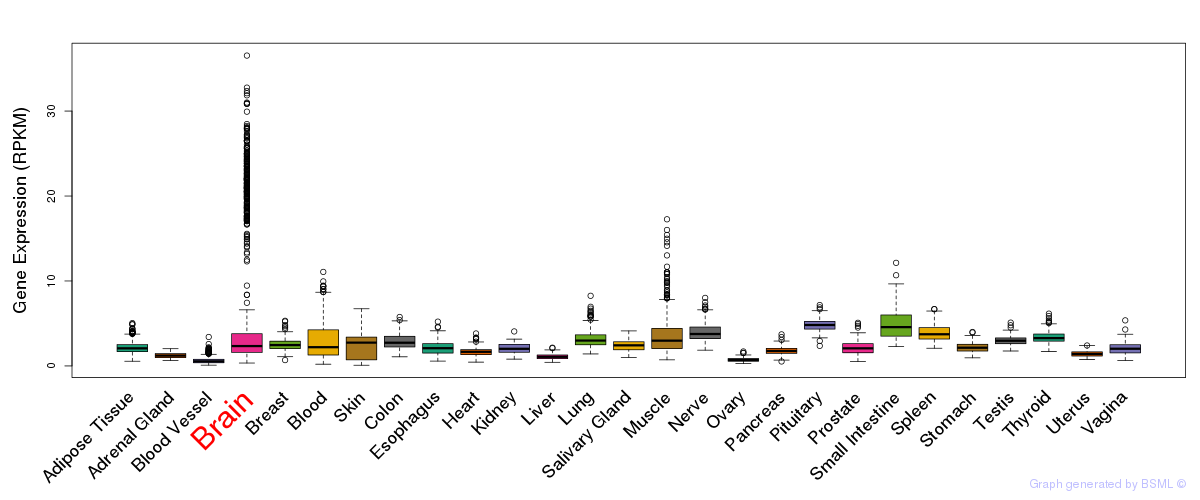
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMA4 | 0.80 | 0.80 |
| PSMA5 | 0.79 | 0.76 |
| SRP19 | 0.78 | 0.77 |
| UBE2T | 0.78 | 0.77 |
| FKBP3 | 0.78 | 0.78 |
| MEA1 | 0.77 | 0.77 |
| ENY2 | 0.76 | 0.75 |
| MED7 | 0.76 | 0.75 |
| TCEAL8 | 0.76 | 0.75 |
| MRPL32 | 0.76 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.66 | -0.58 |
| AF347015.2 | -0.64 | -0.54 |
| AF347015.8 | -0.64 | -0.56 |
| AF347015.15 | -0.64 | -0.57 |
| AF347015.33 | -0.63 | -0.55 |
| MT-CYB | -0.63 | -0.56 |
| MT-CO2 | -0.62 | -0.52 |
| AF347015.27 | -0.60 | -0.55 |
| AF347015.18 | -0.60 | -0.52 |
| AF347015.31 | -0.57 | -0.51 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| LEIN CEREBELLUM MARKERS | 85 | 47 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| LIN TUMOR ESCAPE FROM IMMUNE ATTACK | 18 | 12 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS DN | 46 | 24 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |