Gene Page: AGK
Summary ?
| GeneID | 55750 |
| Symbol | AGK |
| Synonyms | CATC5|CTRCT38|MTDPS10|MULK |
| Description | acylglycerol kinase |
| Reference | MIM:610345|HGNC:HGNC:21869|HPRD:08554| |
| Gene type | protein-coding |
| Map location | 7q34 |
| Pascal p-value | 0.403 |
| Sherlock p-value | 0.869 |
| Fetal beta | -0.249 |
| DMG | 1 (# studies) |
| Support | CELL METABOLISM G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11789684 | 7 | 141251045 | AGK | 5.31E-5 | -0.303 | 0.022 | DMG:Wockner_2014 |
| cg10994314 | 7 | 141251231 | AGK | 4.505E-4 | -0.203 | 0.045 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
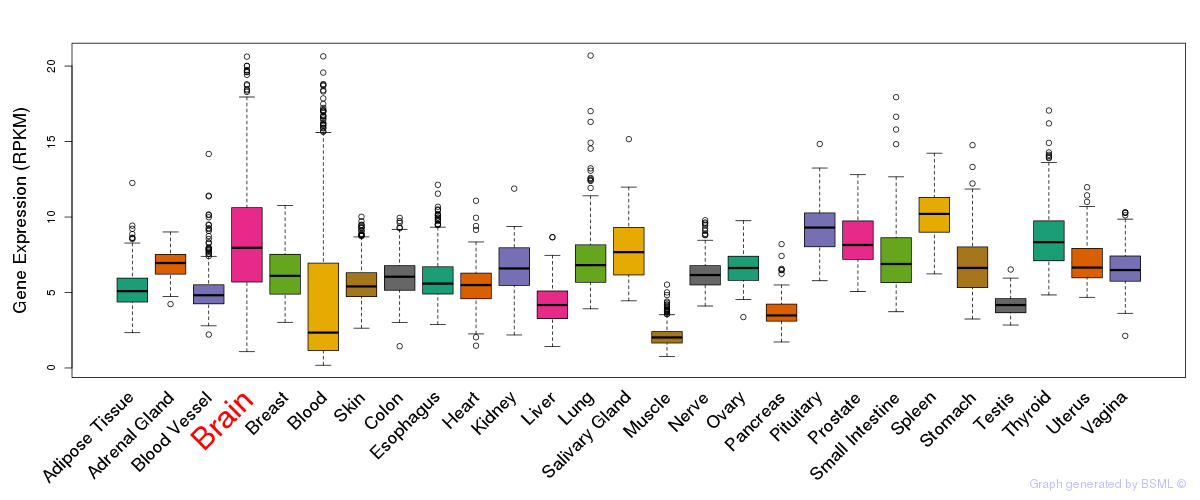
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM171A2 | 0.93 | 0.94 |
| PODXL2 | 0.93 | 0.93 |
| ZNF205 | 0.92 | 0.91 |
| COMTD1 | 0.91 | 0.91 |
| FBL | 0.91 | 0.91 |
| SAMD14 | 0.91 | 0.90 |
| SH2B2 | 0.91 | 0.79 |
| VPS37D | 0.90 | 0.93 |
| GPSM1 | 0.90 | 0.90 |
| PKN1 | 0.90 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.70 | -0.75 |
| AF347015.27 | -0.67 | -0.83 |
| C5orf53 | -0.67 | -0.74 |
| AF347015.33 | -0.66 | -0.83 |
| AF347015.31 | -0.65 | -0.80 |
| S100B | -0.65 | -0.77 |
| MT-CO2 | -0.65 | -0.82 |
| COPZ2 | -0.65 | -0.75 |
| CSRP1 | -0.65 | -0.71 |
| PTGDS | -0.65 | -0.71 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCEROLIPID METABOLISM | 49 | 26 | All SZGR 2.0 genes in this pathway |
| NAM FXYD5 TARGETS DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |