Gene Page: CDK5RAP2
Summary ?
| GeneID | 55755 |
| Symbol | CDK5RAP2 |
| Synonyms | C48|Cep215|MCPH3 |
| Description | CDK5 regulatory subunit associated protein 2 |
| Reference | MIM:608201|HGNC:HGNC:18672|Ensembl:ENSG00000136861|HPRD:09740|Vega:OTTHUMG00000021043 |
| Gene type | protein-coding |
| Map location | 9q33.2 |
| Pascal p-value | 0.084 |
| Sherlock p-value | 0.933 |
| Fetal beta | 0.718 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12576700 | 9 | 123342680 | CDK5RAP2 | -0.024 | 0.39 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs872423 | chr9 | 123149490 | CDK5RAP2 | 55755 | 5.781E-5 | cis | ||
| rs10119527 | chr9 | 123161933 | CDK5RAP2 | 55755 | 7.594E-5 | cis | ||
| rs12379034 | chr9 | 123162665 | CDK5RAP2 | 55755 | 7.766E-6 | cis | ||
| rs12376337 | chr9 | 123162794 | CDK5RAP2 | 55755 | 7.766E-6 | cis | ||
| rs1547266 | chr9 | 123163475 | CDK5RAP2 | 55755 | 1.943E-4 | cis | ||
| rs3780674 | chr9 | 123166918 | CDK5RAP2 | 55755 | 1.943E-4 | cis | ||
| rs16909765 | chr9 | 123172517 | CDK5RAP2 | 55755 | 9.082E-4 | cis | ||
| rs872423 | chr9 | 123149490 | CDK5RAP2 | 55755 | 0.01 | trans | ||
| rs10119527 | chr9 | 123161933 | CDK5RAP2 | 55755 | 0.01 | trans | ||
| rs12379034 | chr9 | 123162665 | CDK5RAP2 | 55755 | 0 | trans | ||
| rs1547266 | chr9 | 123163475 | CDK5RAP2 | 55755 | 0.02 | trans | ||
| rs3780674 | chr9 | 123166918 | CDK5RAP2 | 55755 | 0.02 | trans | ||
| rs16909765 | chr9 | 123172517 | CDK5RAP2 | 55755 | 0.07 | trans | ||
| rs17687831 | chr10 | 103134632 | CDK5RAP2 | 55755 | 0.09 | trans | ||
| rs10883648 | chr10 | 103185180 | CDK5RAP2 | 55755 | 0.09 | trans | ||
| rs10883649 | chr10 | 103186585 | CDK5RAP2 | 55755 | 0.13 | trans | ||
| rs12248356 | chr10 | 103197718 | CDK5RAP2 | 55755 | 0.14 | trans | ||
| rs4436485 | chr10 | 103245360 | CDK5RAP2 | 55755 | 0.07 | trans | ||
| rs3095798 | chr10 | 103299266 | CDK5RAP2 | 55755 | 0.09 | trans | ||
| rs2586047 | chr17 | 55959943 | CDK5RAP2 | 55755 | 0.09 | trans |
Section II. Transcriptome annotation
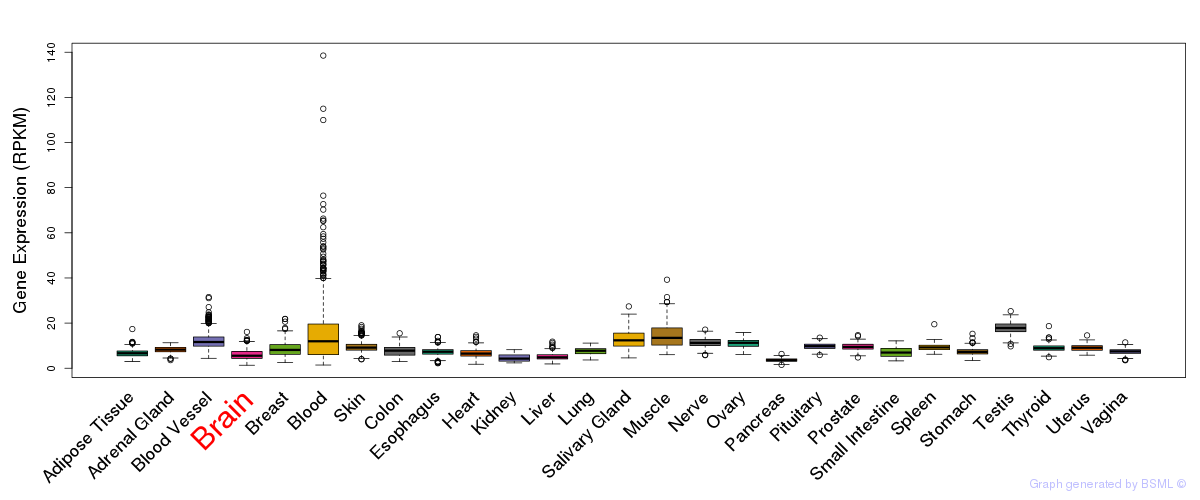
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC4 | 0.80 | 0.81 |
| SLITRK1 | 0.79 | 0.84 |
| TMEM178 | 0.78 | 0.82 |
| ITPK1 | 0.78 | 0.78 |
| LARGE | 0.78 | 0.81 |
| A2BP1 | 0.77 | 0.78 |
| BCAS3 | 0.77 | 0.79 |
| CDH22 | 0.77 | 0.80 |
| SH3GL2 | 0.77 | 0.81 |
| SIPA1L1 | 0.76 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DBI | -0.54 | -0.60 |
| AF347015.21 | -0.49 | -0.42 |
| RAB34 | -0.49 | -0.55 |
| RHOC | -0.49 | -0.53 |
| AP002478.3 | -0.49 | -0.50 |
| PSME2 | -0.48 | -0.50 |
| NSBP1 | -0.47 | -0.51 |
| GNG11 | -0.47 | -0.47 |
| AF347015.18 | -0.46 | -0.42 |
| AL139819.3 | -0.46 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042808 | neuronal Cdc2-like kinase binding | IPI | neuron (GO term level: 7) | 10721722 |
| GO:0008017 | microtubule binding | IDA | 14654843 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | NAS | Brain (GO term level: 7) | 10915792 |
| GO:0045664 | regulation of neuron differentiation | NAS | neuron, neurogenesis (GO term level: 9) | 10721722 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 14654843 | |
| GO:0005856 | cytoskeleton | NAS | 10721722 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCA2 | ABC2 | MGC129761 | ATP-binding cassette, sub-family A (ABC1), member 2 | - | HPRD | 12421765 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C7orf64 | DKFZP564O0523 | DKFZp686D1651 | HSPC304 | chromosome 7 open reading frame 64 | Two-hybrid | BioGRID | 16169070 |
| CDK5 | PSSALRE | cyclin-dependent kinase 5 | Reconstituted Complex | BioGRID | 10915792 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | in vitro in vivo Reconstituted Complex | BioGRID | 10915792 |11882646 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD | 10721722 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| EEF1G | EF1G | GIG35 | eukaryotic translation elongation factor 1 gamma | Two-hybrid | BioGRID | 16169070 |
| HAP1 | HAP2 | HIP5 | HLP | hHLP1 | huntingtin-associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| KAT5 | ESA1 | HTATIP | HTATIP1 | PLIP | TIP | TIP60 | cPLA2 | K(lysine) acetyltransferase 5 | Two-hybrid | BioGRID | 16169070 |
| LUC7L2 | CGI-59 | CGI-74 | FLJ10657 | LUC7B2 | LUC7-like 2 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| MAGEH1 | APR-1 | APR1 | MAGEH | melanoma antigen family H, 1 | Two-hybrid | BioGRID | 16169070 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| SENP2 | AXAM2 | DKFZp762A2316 | KIAA1331 | SMT3IP2 | SUMO1/sentrin/SMT3 specific peptidase 2 | Two-hybrid | BioGRID | 16169070 |
| STARD9 | DKFZp781J069 | FLJ16106 | FLJ21936 | KIAA1300 | StAR-related lipid transfer (START) domain containing 9 | Two-hybrid | BioGRID | 16169070 |
| TBC1D4 | AS160 | DKFZp779C0666 | TBC1 domain family, member 4 | - | HPRD | 12421765 |
| ZNF259 | MGC110983 | ZPR1 | zinc finger protein 259 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE UP | 85 | 57 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER WITH LOH IN CHR9Q | 116 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 6 | 52 | 32 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |