Gene Page: PRKAR2A
Summary ?
| GeneID | 5576 |
| Symbol | PRKAR2A |
| Synonyms | PKR2|PRKAR2 |
| Description | protein kinase cAMP-dependent type II regulatory subunit alpha |
| Reference | MIM:176910|HGNC:HGNC:9391|Ensembl:ENSG00000114302|HPRD:01484|Vega:OTTHUMG00000133540 |
| Gene type | protein-coding |
| Map location | 3p21.3-p21.2 |
| Pascal p-value | 0.021 |
| Fetal beta | 1.025 |
| DMG | 1 (# studies) |
| Support | CANABINOID DOPAMINE INTRACELLULAR SIGNAL TRANSDUCTION SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0096 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22934568 | 3 | 48884614 | PRKAR2A | 4.48E-4 | -0.278 | 0.045 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
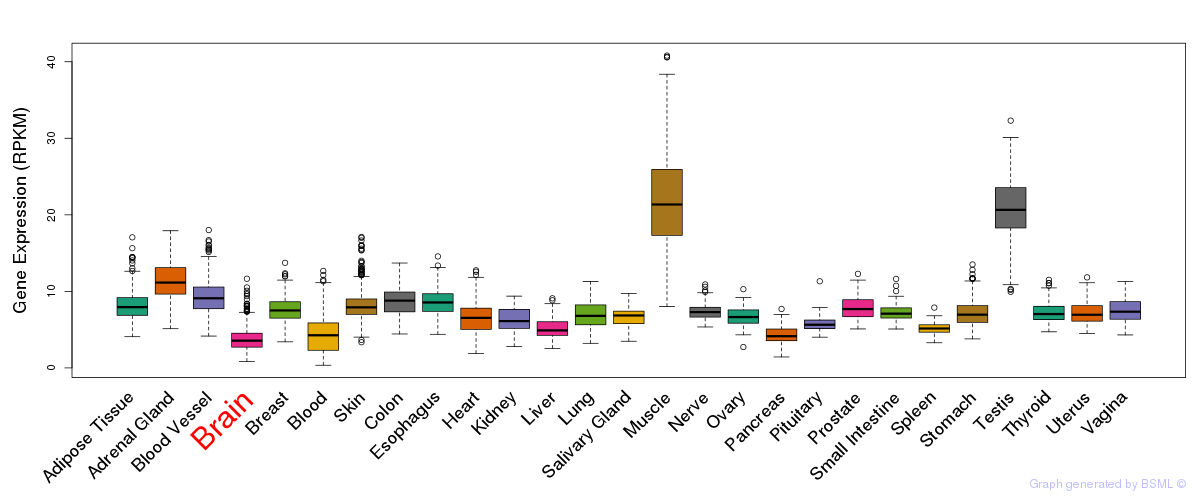
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NCBP1 | 0.90 | 0.89 |
| WDR3 | 0.89 | 0.90 |
| ZMYM4 | 0.89 | 0.90 |
| CTDSPL2 | 0.89 | 0.91 |
| CAND1 | 0.89 | 0.89 |
| SCYL2 | 0.89 | 0.90 |
| KIAA1430 | 0.89 | 0.90 |
| IPO5 | 0.89 | 0.89 |
| NUDT21 | 0.89 | 0.89 |
| VEZT | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.73 | -0.83 |
| AF347015.33 | -0.72 | -0.83 |
| FXYD1 | -0.72 | -0.84 |
| HLA-F | -0.71 | -0.79 |
| AF347015.31 | -0.71 | -0.81 |
| MT-CYB | -0.70 | -0.81 |
| AIFM3 | -0.70 | -0.77 |
| HEPN1 | -0.70 | -0.78 |
| AF347015.8 | -0.70 | -0.82 |
| AF347015.27 | -0.69 | -0.81 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0008603 | cAMP-dependent protein kinase regulator activity | IEA | - | |
| GO:0008603 | cAMP-dependent protein kinase regulator activity | NAS | 9003463 | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0030552 | cAMP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001932 | regulation of protein amino acid phosphorylation | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0009755 | hormone-mediated signaling | EXP | 12626323 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 10799517 | |
| GO:0005737 | cytoplasm | TAS | 10799517 | |
| GO:0005952 | cAMP-dependent protein kinase complex | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 10799517 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | - | HPRD | 9065479 |10764601 |
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | Far Western Reconstituted Complex | BioGRID | 10413680 |10764601 |
| AKAP10 | D-AKAP2 | MGC9414 | PRKA10 | A kinase (PRKA) anchor protein 10 | - | HPRD | 9326583 |
| AKAP11 | AKAP220 | DKFZp781I12161 | FLJ11304 | KIAA0629 | PRKA11 | A kinase (PRKA) anchor protein 11 | - | HPRD,BioGRID | 8621616 |
| AKAP12 | AKAP250 | DKFZp686M0430 | DKFZp686O0331 | FLJ20945 | FLJ97621 | A kinase (PRKA) anchor protein 12 | - | HPRD,BioGRID | 11316952 |
| AKAP13 | AKAP-Lbc | BRX | FLJ11952 | FLJ43341 | HA-3 | Ht31 | LBC | PROTO-LB | PROTO-LBC | c-lbc | A kinase (PRKA) anchor protein 13 | - | HPRD | 1618839 |7929081 |
| AKAP13 | AKAP-Lbc | BRX | FLJ11952 | FLJ43341 | HA-3 | Ht31 | LBC | PROTO-LB | PROTO-LBC | c-lbc | A kinase (PRKA) anchor protein 13 | - | HPRD,BioGRID | 12672969 |
| AKAP14 | AKAP28 | A kinase (PRKA) anchor protein 14 | - | HPRD,BioGRID | 12475942 |
| AKAP2 | AKAPKL | FLJ53197 | PALM2 | PRKA2 | A kinase (PRKA) anchor protein 2 | Far Western Protein-peptide Reconstituted Complex | BioGRID | 9497389 |12672969 |
| AKAP3 | AKAP110 | FSP95 | PRKA3 | SOB1 | A kinase (PRKA) anchor protein 3 | - | HPRD,BioGRID | 10319321 |11278869 |11696326 |
| AKAP4 | AKAP82 | FSC1 | HI | hAKAP82 | p82 | A kinase (PRKA) anchor protein 4 | Reconstituted Complex | BioGRID | 9852104 |
| AKAP5 | AKAP75 | AKAP79 | H21 | A kinase (PRKA) anchor protein 5 | - | HPRD,BioGRID | 12672969 |
| AKAP6 | ADAP100 | ADAP6 | AKAP100 | KIAA0311 | MGC165020 | PRKA6 | mAKAP | A kinase (PRKA) anchor protein 6 | - | HPRD | 10413680 |
| AKAP6 | ADAP100 | ADAP6 | AKAP100 | KIAA0311 | MGC165020 | PRKA6 | mAKAP | A kinase (PRKA) anchor protein 6 | Protein-peptide | BioGRID | 12672969 |
| AKAP7 | AKAP18 | A kinase (PRKA) anchor protein 7 | - | HPRD,BioGRID | 12672969 |
| AKAP8 | AKAP95 | DKFZp586B1222 | A kinase (PRKA) anchor protein 8 | - | HPRD,BioGRID | 9473338 |10601332 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD,BioGRID | 10358086 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | - | HPRD | 10358086 |11799244 |
| ARFGEF2 | BIG2 | FLJ23723 | dJ1164I10.1 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) | Reconstituted Complex Two-hybrid | BioGRID | 12571360 |
| CBFA2T3 | ETO2 | MTG16 | MTGR2 | ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | - | HPRD,BioGRID | 11823486 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | PKA II interacts with and phosphorylates CFTR. | BIND | 10799517 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | PKA II interacts with Ezrin. | BIND | 9009265 |10799517 |12080047 |
| GNA13 | G13 | MGC46138 | guanine nucleotide binding protein (G protein), alpha 13 | Affinity Capture-Western | BioGRID | 11696326 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-Western | BioGRID | 12147701 |
| KCNQ1 | ATFB1 | FLJ26167 | JLNS1 | KCNA8 | KCNA9 | KVLQT1 | Kv1.9 | Kv7.1 | LQT | LQT1 | RWS | SQT2 | WRS | potassium voltage-gated channel, KQT-like subfamily, member 1 | Affinity Capture-Western | BioGRID | 11799244 |
| MYCBPAP | AMAP-1 | AMAP1 | DKFZp434N1415 | MYCBP associated protein | - | HPRD | 12151104 |
| NBEA | BCL8B | LYST2 | neurobeachin | - | HPRD | 11102458 |
| PALM2-AKAP2 | AKAP2 | DKFZp686H1948 | PALM2-AKAP2 readthrough transcript | - | HPRD | 9497389 |12672969 |
| PDE4A | DPDE2 | PDE4 | PDE46 | phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) | Affinity Capture-Western | BioGRID | 11296225 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | PKA forms a dimer. This interaction was modeled on a demonstrated interaction between mouse PKA (PDB ID: 1L6E_B) and mouse PKA (PDB ID: 1L6E_A). | BIND | 9295304 |10074940 |11985580 |
| RAB32 | - | RAB32, member RAS oncogene family | Far Western Protein-peptide Two-hybrid | BioGRID | 12186851 |12672969 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD,BioGRID | 11593431 |
| RYR2 | ARVC2 | ARVD2 | VTSIP | ryanodine receptor 2 (cardiac) | - | HPRD,BioGRID | 10830164 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | WAVE-1 interacts with PKA. | BIND | 10970852 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CSK PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP95 PATHWAY | 12 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGPCR PATHWAY | 13 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CFTR PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GATA3 PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1R PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PLCE PATHWAY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP13 PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STATHMIN PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM1 PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4 | 61 | 34 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM HIGH RISK UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-149 | 842 | 848 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-15/16/195/424/497 | 341 | 348 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-185 | 278 | 285 | 1A,m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-378* | 307 | 314 | 1A,m8 | hsa-miR-422b | CUGGACUUGGAGUCAGAAGGCC |
| hsa-miR-422a | CUGGACUUAGGGUCAGAAGGCC | ||||
| miR-503 | 342 | 348 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-504 | 846 | 852 | m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.