Gene Page: ST6GALNAC1
Summary ?
| GeneID | 55808 |
| Symbol | ST6GALNAC1 |
| Synonyms | HSY11339|SIAT7A|ST6GalNAcI|STYI |
| Description | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 |
| Reference | MIM:610138|HGNC:HGNC:23614|Ensembl:ENSG00000070526|HPRD:15340|Vega:OTTHUMG00000180369 |
| Gene type | protein-coding |
| Map location | 17q25.1 |
| Pascal p-value | 0.28 |
| Fetal beta | -0.57 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
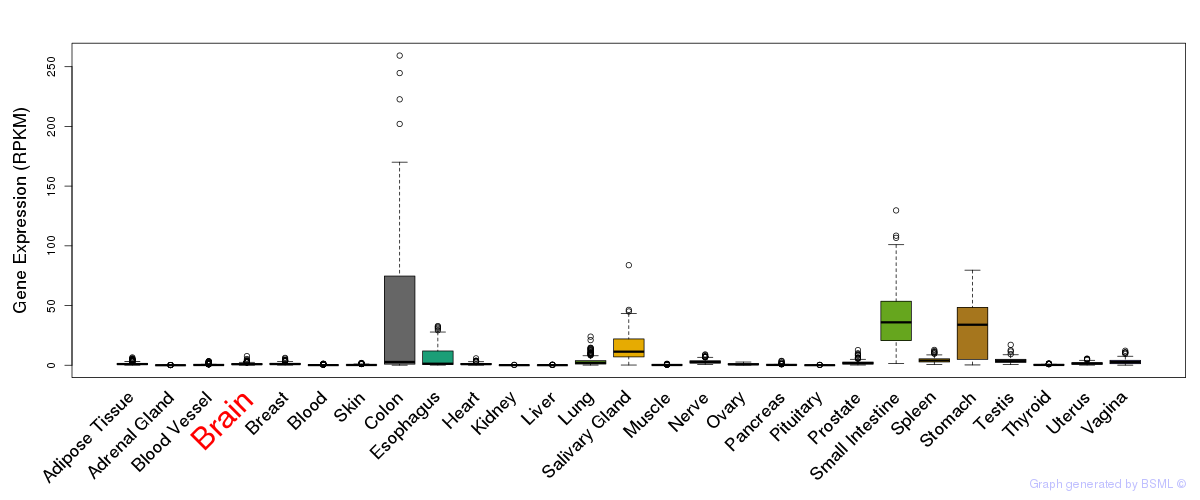
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM49 | 0.88 | 0.86 |
| GFM2 | 0.88 | 0.90 |
| CRBN | 0.87 | 0.87 |
| ORC5L | 0.87 | 0.85 |
| ERGIC2 | 0.86 | 0.86 |
| PNPLA8 | 0.86 | 0.85 |
| PMPCB | 0.86 | 0.82 |
| CCNC | 0.85 | 0.82 |
| YPEL5 | 0.85 | 0.84 |
| ARL6IP5 | 0.85 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.58 | -0.54 |
| AF347015.2 | -0.56 | -0.47 |
| AF347015.26 | -0.55 | -0.47 |
| AF347015.8 | -0.55 | -0.47 |
| MT-CO2 | -0.53 | -0.46 |
| MT-CYB | -0.53 | -0.46 |
| AF347015.21 | -0.53 | -0.41 |
| AF347015.15 | -0.53 | -0.45 |
| GPT | -0.51 | -0.52 |
| AC100783.1 | -0.51 | -0.54 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG O GLYCAN BIOSYNTHESIS | 30 | 16 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR UP | 85 | 54 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| MCCOLLUM GELDANAMYCIN RESISTANCE UP | 10 | 9 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 DN | 120 | 71 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 5 DN | 50 | 31 | All SZGR 2.0 genes in this pathway |