Gene Page: ZNF302
Summary ?
| GeneID | 55900 |
| Symbol | ZNF302 |
| Synonyms | HSD16|MST154|MSTP154|ZNF135L|ZNF140L|ZNF327 |
| Description | zinc finger protein 302 |
| Reference | HGNC:HGNC:13848|Ensembl:ENSG00000089335|HPRD:15765|Vega:OTTHUMG00000163329 |
| Gene type | protein-coding |
| Map location | 19q13.11 |
| Pascal p-value | 0.362 |
| Sherlock p-value | 0.658 |
| Fetal beta | 0.941 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg19850565 | 19 | 35168394 | ZNF302 | 1.58E-9 | -0.009 | 1.45E-6 | DMG:Jaffe_2016 |
| cg13337697 | 19 | 35168649 | ZNF302 | 3.94E-9 | -0.013 | 2.44E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8112651 | 19 | 35135509 | ZNF302 | ENSG00000089335.16 | 1.13E-7 | 0.01 | -33043 | gtex_brain_putamen_basal |
| rs10426406 | 19 | 35139955 | ZNF302 | ENSG00000089335.16 | 1.128E-7 | 0.01 | -28597 | gtex_brain_putamen_basal |
| rs12984680 | 19 | 35146785 | ZNF302 | ENSG00000089335.16 | 1.07E-7 | 0.01 | -21767 | gtex_brain_putamen_basal |
| rs28871304 | 19 | 35149001 | ZNF302 | ENSG00000089335.16 | 1.068E-7 | 0.01 | -19551 | gtex_brain_putamen_basal |
| rs12982639 | 19 | 35152015 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -16537 | gtex_brain_putamen_basal |
| rs12982680 | 19 | 35152081 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -16471 | gtex_brain_putamen_basal |
| rs12986415 | 19 | 35153457 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -15095 | gtex_brain_putamen_basal |
| rs200722278 | 19 | 35153864 | ZNF302 | ENSG00000089335.16 | 7.526E-7 | 0.01 | -14688 | gtex_brain_putamen_basal |
| rs35140779 | 19 | 35153867 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -14685 | gtex_brain_putamen_basal |
| rs34242118 | 19 | 35155913 | ZNF302 | ENSG00000089335.16 | 7.359E-7 | 0.01 | -12639 | gtex_brain_putamen_basal |
| rs28605044 | 19 | 35156434 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -12118 | gtex_brain_putamen_basal |
| rs73043937 | 19 | 35158535 | ZNF302 | ENSG00000089335.16 | 4.636E-7 | 0.01 | -10017 | gtex_brain_putamen_basal |
| rs10411637 | 19 | 35158848 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -9704 | gtex_brain_putamen_basal |
| rs28449888 | 19 | 35165633 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -2919 | gtex_brain_putamen_basal |
| rs10413838 | 19 | 35167690 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | -862 | gtex_brain_putamen_basal |
| rs8107495 | 19 | 35168545 | ZNF302 | ENSG00000089335.16 | 1.644E-6 | 0.01 | -7 | gtex_brain_putamen_basal |
| rs8106282 | 19 | 35168629 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | 77 | gtex_brain_putamen_basal |
| rs34893432 | 19 | 35168964 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | 412 | gtex_brain_putamen_basal |
| rs12977444 | 19 | 35169231 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 679 | gtex_brain_putamen_basal |
| rs10421533 | 19 | 35169468 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 916 | gtex_brain_putamen_basal |
| rs8104524 | 19 | 35171287 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 2735 | gtex_brain_putamen_basal |
| rs34679438 | 19 | 35173003 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 4451 | gtex_brain_putamen_basal |
| rs10410491 | 19 | 35173452 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 4900 | gtex_brain_putamen_basal |
| rs66509108 | 19 | 35174227 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 5675 | gtex_brain_putamen_basal |
| rs11547623 | 19 | 35174693 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | 6141 | gtex_brain_putamen_basal |
| rs10401309 | 19 | 35175749 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | 7197 | gtex_brain_putamen_basal |
| rs7249792 | 19 | 35176752 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 8200 | gtex_brain_putamen_basal |
| rs376995651 | 19 | 35177316 | ZNF302 | ENSG00000089335.16 | 1.066E-7 | 0.01 | 8764 | gtex_brain_putamen_basal |
| rs10423377 | 19 | 35178369 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 9817 | gtex_brain_putamen_basal |
| rs10424028 | 19 | 35178619 | ZNF302 | ENSG00000089335.16 | 1.622E-6 | 0.01 | 10067 | gtex_brain_putamen_basal |
| rs34050875 | 19 | 35179623 | ZNF302 | ENSG00000089335.16 | 1.624E-6 | 0.01 | 11071 | gtex_brain_putamen_basal |
| rs28628519 | 19 | 35180035 | ZNF302 | ENSG00000089335.16 | 1.624E-6 | 0.01 | 11483 | gtex_brain_putamen_basal |
| rs74183313 | 19 | 35180199 | ZNF302 | ENSG00000089335.16 | 1.624E-6 | 0.01 | 11647 | gtex_brain_putamen_basal |
| rs28446844 | 19 | 35180798 | ZNF302 | ENSG00000089335.16 | 1.624E-6 | 0.01 | 12246 | gtex_brain_putamen_basal |
| rs17626118 | 19 | 35181403 | ZNF302 | ENSG00000089335.16 | 1.626E-6 | 0.01 | 12851 | gtex_brain_putamen_basal |
| rs10418293 | 19 | 35181828 | ZNF302 | ENSG00000089335.16 | 1.14E-7 | 0.01 | 13276 | gtex_brain_putamen_basal |
| rs10426964 | 19 | 35183476 | ZNF302 | ENSG00000089335.16 | 1.316E-7 | 0.01 | 14924 | gtex_brain_putamen_basal |
| rs28462181 | 19 | 35205960 | ZNF302 | ENSG00000089335.16 | 1.517E-6 | 0.01 | 37408 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
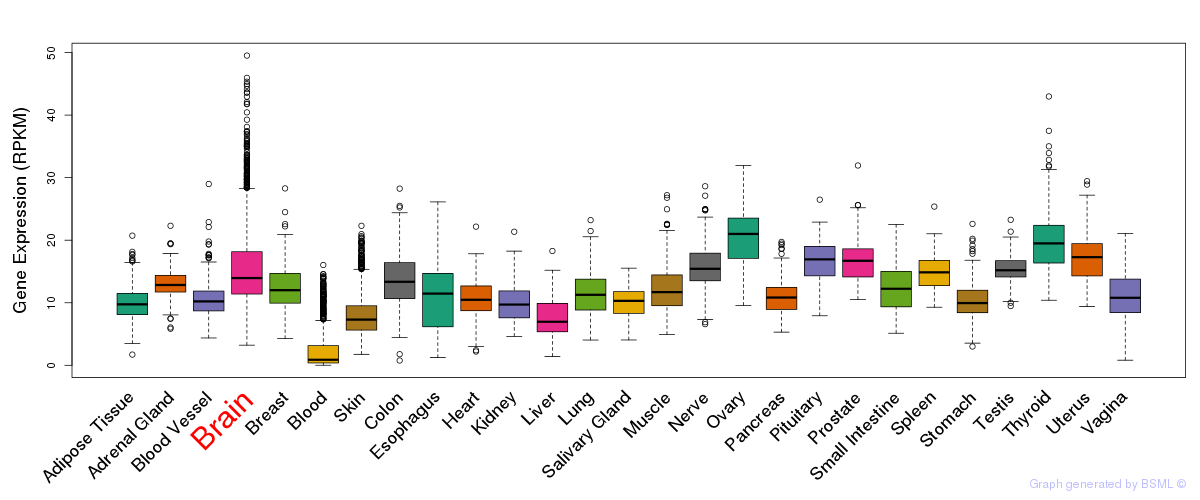
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS RESPONSIVE TO ESTROGEN UP | 31 | 20 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |