Gene Page: MAPK1
Summary ?
| GeneID | 5594 |
| Symbol | MAPK1 |
| Synonyms | ERK|ERK-2|ERK2|ERT1|MAPK2|P42MAPK|PRKM1|PRKM2|p38|p40|p41|p41mapk|p42-MAPK |
| Description | mitogen-activated protein kinase 1 |
| Reference | MIM:176948|HGNC:HGNC:6871|Ensembl:ENSG00000100030|HPRD:01496|Vega:OTTHUMG00000030508 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.481 |
| Fetal beta | -1.658 |
| eGene | Myers' cis & trans |
| Support | CANABINOID INTRACELLULAR SIGNAL TRANSDUCTION METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1451 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10504300 | chr8 | 60714730 | MAPK1 | 5594 | 0.12 | trans | ||
| rs493713 | chr8 | 60882998 | MAPK1 | 5594 | 0.16 | trans |
Section II. Transcriptome annotation
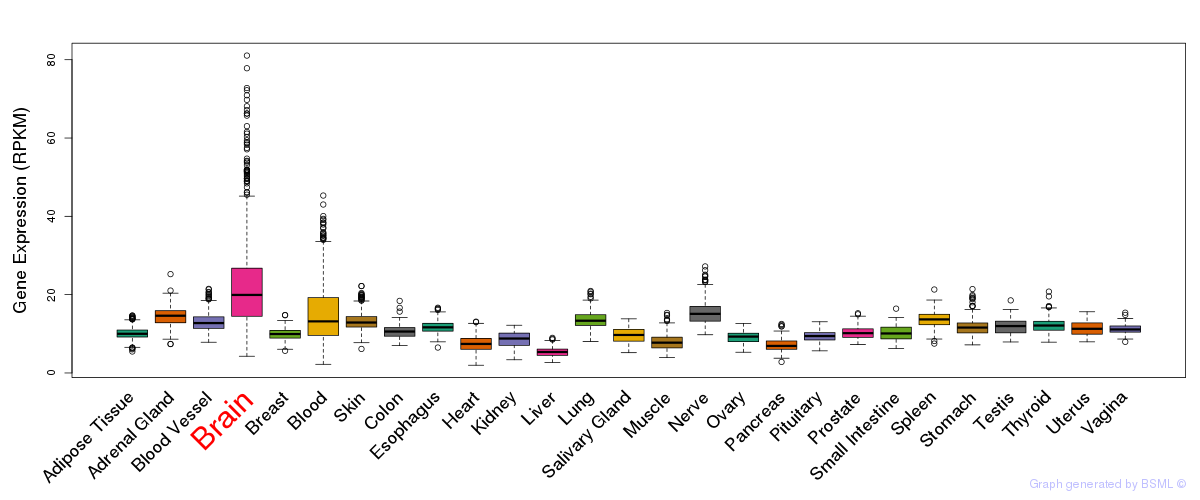
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004707 | MAP kinase activity | TAS | 10706854 | |
| GO:0001784 | phosphotyrosine binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9632734 |16286470 |16291755 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | EXP | 16626623 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 10051431 |
| GO:0000165 | MAPKKK cascade | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0006917 | induction of apoptosis | TAS | 10958679 | |
| GO:0009887 | organ morphogenesis | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0019858 | cytosine metabolic process | IEA | - | |
| GO:0006974 | response to DNA damage stimulus | IEA | - | |
| GO:0006935 | chemotaxis | TAS | 10706854 | |
| GO:0032496 | response to lipopolysaccharide | IEA | - | |
| GO:0031663 | lipopolysaccharide-mediated signaling pathway | IEA | - | |
| GO:0043330 | response to exogenous dsRNA | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0045596 | negative regulation of cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10066798 |10878576 | |
| GO:0005856 | cytoskeleton | IDA | 18029348 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 9687510 |16456541 |16626623 |17322878 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAM17 | CD156b | MGC71942 | TACE | cSVP | ADAM metallopeptidase domain 17 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 12058067 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Reconstituted Complex | BioGRID | 10318905 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | Biochemical Activity | BioGRID | 7737129 |
| ATP1A1 | MGC3285 | MGC51750 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | Biochemical Activity | BioGRID | 15069082 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 15225643 |
| C1QBP | GC1QBP | HABP1 | SF2p32 | gC1Q-R | gC1qR | p32 | complement component 1, q subcomponent binding protein | - | HPRD,BioGRID | 11866440 |
| CALCOCO1 | Cocoa | KIAA1536 | PP13275 | calphoglin | calcium binding and coiled-coil domain 1 | KIAA1536 is phosphorylated by MAPK1. This interaction was modeled on a demonstrated interaction between human KIAA1536 and MAPK1 from an unspecified species. | BIND | 15489336 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 12792650 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | Biochemical Activity | BioGRID | 11500490 |
| DAPK1 | DAPK | DKFZp781I035 | death-associated protein kinase 1 | DAPK interacts with Erk2. | BIND | 15616583 |
| DUSP1 | CL100 | HVH1 | MKP-1 | MKP1 | PTPN10 | dual specificity phosphatase 1 | - | HPRD,BioGRID | 11278799 |
| DUSP16 | KIAA1700 | MGC129701 | MGC129702 | MKP-7 | MKP7 | dual specificity phosphatase 16 | - | HPRD,BioGRID | 11489891 |
| DUSP22 | JKAP | JSP1 | MKPX | VHX | dual specificity phosphatase 22 | Affinity Capture-Western Biochemical Activity | BioGRID | 11346645 |
| DUSP3 | VHR | dual specificity phosphatase 3 | - | HPRD,BioGRID | 10224087 |
| DUSP4 | HVH2 | MKP-2 | MKP2 | TYP | dual specificity phosphatase 4 | - | HPRD | 7535768 |
| DUSP6 | MKP3 | PYST1 | dual specificity phosphatase 6 | - | HPRD,BioGRID | 9535927 |
| DUSP6 | MKP3 | PYST1 | dual specificity phosphatase 6 | - | HPRD | 9535927 |15269220 |
| DYRK1B | MIRK | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B | - | HPRD,BioGRID | 10910078 |
| ELK1 | - | ELK1, member of ETS oncogene family | Mapk1 (p42Mapk) phosphorylates Elk1 at Ser383. | BIND | 15782123 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD | 8208531 |8586671 |
| ELK1 | - | ELK1, member of ETS oncogene family | Biochemical Activity Reconstituted Complex | BioGRID | 8586671 |12594221 |
| EPOR | MGC138358 | erythropoietin receptor | - | HPRD | 12538595 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 12093745 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14729955 |
| FHL3 | MGC19547 | MGC23614 | MGC8696 | SLIM2 | four and a half LIM domains 3 | - | HPRD | 14729955 |
| GATA4 | MGC126629 | GATA binding protein 4 | Biochemical Activity | BioGRID | 11585926 |
| GMFB | GMF | glia maturation factor, beta | - | HPRD | 8639570 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | - | HPRD,BioGRID | 11114188 |
| HMMR | CD168 | IHABP | MGC119494 | MGC119495 | RHAMM | hyaluronan-mediated motility receptor (RHAMM) | Affinity Capture-Western | BioGRID | 11403955 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 11748628 |
| IER3 | DIF-2 | DIF2 | GLY96 | IEX-1 | IEX-1L | IEX1 | PRG1 | immediate early response 3 | Biochemical Activity | BioGRID | 12356731 |
| IFNAR1 | AVP | IFN-alpha-REC | IFNAR | IFNBR | IFRC | interferon (alpha, beta and omega) receptor 1 | - | HPRD | 9029147 |
| INSR | CD220 | HHF5 | insulin receptor | Affinity Capture-Western | BioGRID | 11409918 |
| KLF11 | FKLF | FKLF1 | MODY7 | TIEG2 | Tieg3 | Kruppel-like factor 11 | - | HPRD | 12006497 |
| KSR2 | FLJ25965 | kinase suppressor of ras 2 | - | HPRD | 12975377 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | MEK1 interacts with and phosphorylates ERK2. This interaction is modeled on a demonstrated interaction between human MEK1 and rat ERK2. | BIND | 11134045 |
| MAP2K1 | MAPKK1 | MEK1 | MKK1 | PRKMK1 | mitogen-activated protein kinase kinase 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10757792 |11266467 |11279118 |11352917 |11823456 |12697810 |
| MAP2K2 | FLJ26075 | MAPKK2 | MEK2 | MKK2 | PRKMK2 | mitogen-activated protein kinase kinase 2 | MEK2 interacts with ERK2. This interaction is modeled on a demonstrated interaction between human MEK2 and rat ERK2. | BIND | 11134045 |
| MAP2K2 | FLJ26075 | MAPKK2 | MEK2 | MKK2 | PRKMK2 | mitogen-activated protein kinase kinase 2 | - | HPRD,BioGRID | 11823456 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western | BioGRID | 10969079 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | MEKK1 interacts with ERK2. | BIND | 10969079 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | - | HPRD,BioGRID | 12697810 |
| MAPKAPK2 | MK2 | mitogen-activated protein kinase-activated protein kinase 2 | Affinity Capture-Western | BioGRID | 15094067 |
| MAPKAPK5 | PRAK | mitogen-activated protein kinase-activated protein kinase 5 | - | HPRD | 9628874 |
| MAPKSP1 | MAP2K1IP1 | MAPBP | MP1 | MAPK scaffold protein 1 | - | HPRD,BioGRID | 11266467 |
| MBP | MGC99675 | myelin basic protein | MBP interacts with MAPK1. This interaction was modeled on a demonstrated interaction between MBP from an unspecified species, and monkey and mouse MAPK1. | BIND | 9792705 |
| MKNK1 | MNK1 | MAP kinase interacting serine/threonine kinase 1 | Reconstituted Complex Two-hybrid | BioGRID | 9155017 |
| MKNK2 | GPRK7 | MNK2 | MAP kinase interacting serine/threonine kinase 2 | - | HPRD,BioGRID | 9155017 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Biochemical Activity Reconstituted Complex | BioGRID | 7957875 |9207092 |15210690 |
| NEK2 | HsPK21 | NEK2A | NLK1 | NIMA (never in mitosis gene a)-related kinase 2 | - | HPRD,BioGRID | 15358203 |
| PEA15 | HMAT1 | HUMMAT1H | MAT1 | MAT1H | PEA-15 | PED | phosphoprotein enriched in astrocytes 15 | - | HPRD,BioGRID | 11702783 |
| PEBP1 | HCNP | PBP | PEBP | RKIP | phosphatidylethanolamine binding protein 1 | - | HPRD,BioGRID | 10757792 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | Affinity Capture-Western | BioGRID | 10801826 |
| PLAT | DKFZp686I03148 | T-PA | TPA | plasminogen activator, tissue | - | HPRD | 15861134 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | Affinity Capture-Western | BioGRID | 11350735 |
| PTPN5 | FLJ14427 | PTPSTEP | STEP | protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) | Reconstituted Complex | BioGRID | 9857190 |
| PTPN7 | BPTP-4 | HEPTP | LC-PTP | LPTP | PTPNI | protein tyrosine phosphatase, non-receptor type 7 | - | HPRD,BioGRID | 10206983 |10702794 |
| PTPRH | FLJ39938 | MGC133058 | MGC133059 | SAP-1 | protein tyrosine phosphatase, receptor type, H | - | HPRD | 11278335 |
| PTPRR | DKFZp781C1038 | EC-PTP | FLJ34328 | MGC131968 | MGC148170 | PCPTP1 | PTP-SL | PTPBR7 | PTPRQ | protein tyrosine phosphatase, receptor type, R | - | HPRD,BioGRID | 9857190 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 11409918 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf-1 interacts with ERK2. This interaction was modelled on a demonstrated interaction between human Raf-1 and ERK2 from an unspecified species. | BIND | 15664191 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | RB1 is phosphorylated by MAPK1. This interaction was modeled on a demonstrated interaction between human KIAA1536 and MAPK1 from an unspecified species. | BIND | 15489336 |
| RPS6KA1 | HU-1 | MAPKAPK1A | RSK | RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 | - | HPRD,BioGRID | 9915826 |12594221 |12832467 |
| RPS6KA2 | HU-2 | MAPKAPK1C | RSK | RSK3 | S6K-alpha | S6K-alpha2 | p90-RSK3 | pp90RSK3 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | Rsk3 interacts with ERK2. This interaction was modelled on a demonstrated interaction between human Rsk3 and monkey ERK2. | BIND | 9915826 |
| RPS6KA2 | HU-2 | MAPKAPK1C | RSK | RSK3 | S6K-alpha | S6K-alpha2 | p90-RSK3 | pp90RSK3 | ribosomal protein S6 kinase, 90kDa, polypeptide 2 | - | HPRD,BioGRID | 8939914 |
| RPS6KA3 | CLS | HU-3 | ISPK-1 | MAPKAPK1B | MRX19 | RSK | RSK2 | S6K-alpha3 | p90-RSK2 | pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 | Affinity Capture-Western | BioGRID | 8939914 |9915826 |
| RPS6KA3 | CLS | HU-3 | ISPK-1 | MAPKAPK1B | MRX19 | RSK | RSK2 | S6K-alpha3 | p90-RSK2 | pp90RSK2 | ribosomal protein S6 kinase, 90kDa, polypeptide 3 | - | HPRD | 12832467 |
| RPS6KA4 | MSK2 | RSK-B | ribosomal protein S6 kinase, 90kDa, polypeptide 4 | - | HPRD | 9687510 |
| RPS6KA5 | MGC1911 | MSK1 | MSPK1 | RLPK | ribosomal protein S6 kinase, 90kDa, polypeptide 5 | Phenotypic Enhancement | BioGRID | 9687510 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 10996427 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | Affinity Capture-Western | BioGRID | 12121974 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD | 11279280 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | Affinity Capture-Western | BioGRID | 12121974 |
| SORBS3 | SCAM-1 | SCAM1 | SH3D4 | VINEXIN | sorbin and SH3 domain containing 3 | Affinity Capture-Western Biochemical Activity | BioGRID | 15184391 |
| SREBF1 | SREBP-1c | SREBP1 | bHLHd1 | sterol regulatory element binding transcription factor 1 | Biochemical Activity | BioGRID | 10627507 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | - | HPRD,BioGRID | 10627507 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 10194762 |10996427 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | - | HPRD | 10996427 |
| TH | TYH | tyrosine hydroxylase | Biochemical Activity | BioGRID | 1347949 |
| TNFSF11 | CD254 | ODF | OPGL | OPTB2 | RANKL | TRANCE | hRANKL2 | sOdf | tumor necrosis factor (ligand) superfamily, member 11 | - | HPRD | 10635328 |
| TNIP1 | ABIN-1 | KIAA0113 | NAF1 | VAN | TNFAIP3 interacting protein 1 | Naf1-alpha interacts with ERK2. | BIND | 12220502 |
| TNIP1 | ABIN-1 | KIAA0113 | NAF1 | VAN | TNFAIP3 interacting protein 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12220502 |
| TNKS2 | PARP-5b | PARP-5c | PARP5B | PARP5C | TANK2 | TNKL | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | Biochemical Activity | BioGRID | 10988299 |
| TOB1 | APRO6 | MGC104792 | MGC34446 | PIG49 | TOB | TROB | TROB1 | transducer of ERBB2, 1 | - | HPRD,BioGRID | 12151396 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 10958792 |
| TPR | - | translocated promoter region (to activated MET oncogene) | - | HPRD,BioGRID | 12594221 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15851026 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Erk2 interacts with TSC2. This interaction was modelled on a demonstrated interaction between Erk2 from an unspecified species and human TSC2. | BIND | 15851026 |
| UBR5 | DD5 | EDD | EDD1 | FLJ11310 | HYD | KIAA0896 | MGC57263 | ubiquitin protein ligase E3 component n-recognin 5 | - | HPRD,BioGRID | 12594221 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 8900182 |9013873 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Vav interacts with ERK2. | BIND | 9013873 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG ALDOSTERONE REGULATED SODIUM REABSORPTION | 42 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPPA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDMAC PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CERAMIDE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CXCR4 PATHWAY | 24 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GH PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HCMV PATHWAY | 17 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PYK2 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1R PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARENRF2 PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EDG1 PATHWAY | 27 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDK5 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BAD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EIF4 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK5 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAL PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARR MAPK PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN SRC PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPRY PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| SIG CD40PATHWAYMAP | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA S PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| SIG IL4RECEPTOR IN B LYPHOCYTES | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| SA PTEN PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P4 PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID AR NONGENOMIC PATHWAY | 31 | 27 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID ANTHRAX PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| PID LYMPH ANGIOGENESIS PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SPRY REGULATION OF FGF SIGNALING | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | 22 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | 20 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME ARMS MEDIATED ACTIVATION | 17 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLONGED ERK ACTIVATION EVENTS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO P38 VIA RIT AND RIN | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERKS ARE INACTIVATED | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC1 EVENTS IN EGFR SIGNALING | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME FRS2 MEDIATED CASCADE | 36 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME RECYCLING PATHWAY OF L1 | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 2 SIGNALING | 41 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME SOS MEDIATED SIGNALLING | 14 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME RAF MAP KINASE CASCADE | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC MEDIATED SIGNALLING | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC RELATED EVENTS | 17 | 13 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 MCF10A | 43 | 26 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN D6MIT150 QTL TRANS | 9 | 7 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| GAZIN EPIGENETIC SILENCING BY KRAS | 26 | 16 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| HUNSBERGER EXERCISE REGULATED GENES | 31 | 26 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 3882 | 3888 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-132/212 | 1379 | 1386 | 1A,m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-181 | 3842 | 3848 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-186 | 3798 | 3804 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-204/211 | 3259 | 3265 | 1A | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-214 | 1168 | 1174 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-217 | 305 | 311 | m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-320 | 652 | 658 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-326 | 184 | 190 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
| miR-330 | 3194 | 3200 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-34b | 1594 | 1600 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-409-5p | 964 | 970 | 1A | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-493-5p | 1642 | 1648 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-539 | 1530 | 1537 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-543 | 3843 | 3849 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.