Gene Page: SEPT3
Summary ?
| GeneID | 55964 |
| Symbol | SEPT3 |
| Synonyms | SEP3|bK250D10.3 |
| Description | septin 3 |
| Reference | MIM:608314|HGNC:HGNC:10750|Ensembl:ENSG00000100167|HPRD:10515|Vega:OTTHUMG00000030494 |
| Gene type | protein-coding |
| Map location | 22q13.2 |
| Pascal p-value | 7.165E-9 |
| Sherlock p-value | 0.719 |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
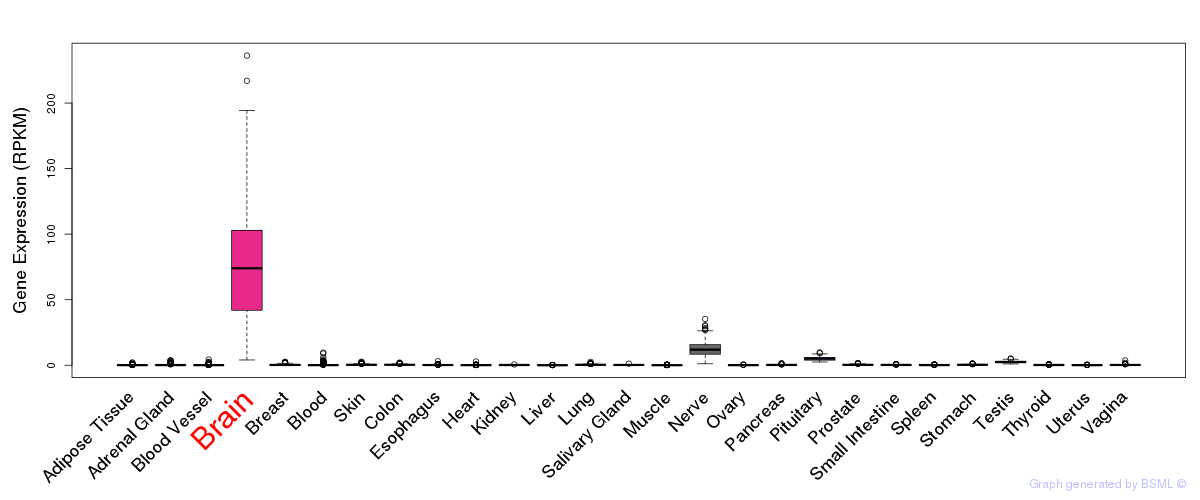
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003674 | molecular_function | ND | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005525 | GTP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000910 | cytokinesis | NAS | - | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031105 | septin complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 49 | 55 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-137 | 760 | 766 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-181 | 914 | 920 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-203.1 | 6 | 12 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-224 | 1025 | 1031 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-23 | 824 | 830 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 823 | 830 | 1A,m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-324-5p | 1085 | 1092 | 1A,m8 | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-328 | 55 | 62 | 1A,m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-34/449 | 938 | 944 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-342 | 439 | 445 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-493-5p | 1224 | 1230 | 1A | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-543 | 915 | 921 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.