Gene Page: AJAP1
Summary ?
| GeneID | 55966 |
| Symbol | AJAP1 |
| Synonyms | MOT8|SHREW-1|SHREW1 |
| Description | adherens junctions associated protein 1 |
| Reference | MIM:610972|HGNC:HGNC:30801|Ensembl:ENSG00000196581|HPRD:15339|Vega:OTTHUMG00000000645 |
| Gene type | protein-coding |
| Map location | 1p36.32 |
| Pascal p-value | 0.213 |
| Fetal beta | -1.239 |
| DMG | 1 (# studies) |
| eGene | Hypothalamus Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26662102 | 1 | 4758079 | AJAP1 | 5.74E-5 | 0.486 | 0.022 | DMG:Wockner_2014 |
| cg06966242 | 1 | 4832432 | AJAP1 | 3.307E-4 | 0.553 | 0.041 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2806023 | chrX | 95245934 | AJAP1 | 55966 | 0.01 | trans | ||
| rs2806022 | chrX | 95246284 | AJAP1 | 55966 | 0.01 | trans | ||
| rs2806021 | chrX | 95246404 | AJAP1 | 55966 | 0.02 | trans |
Section II. Transcriptome annotation
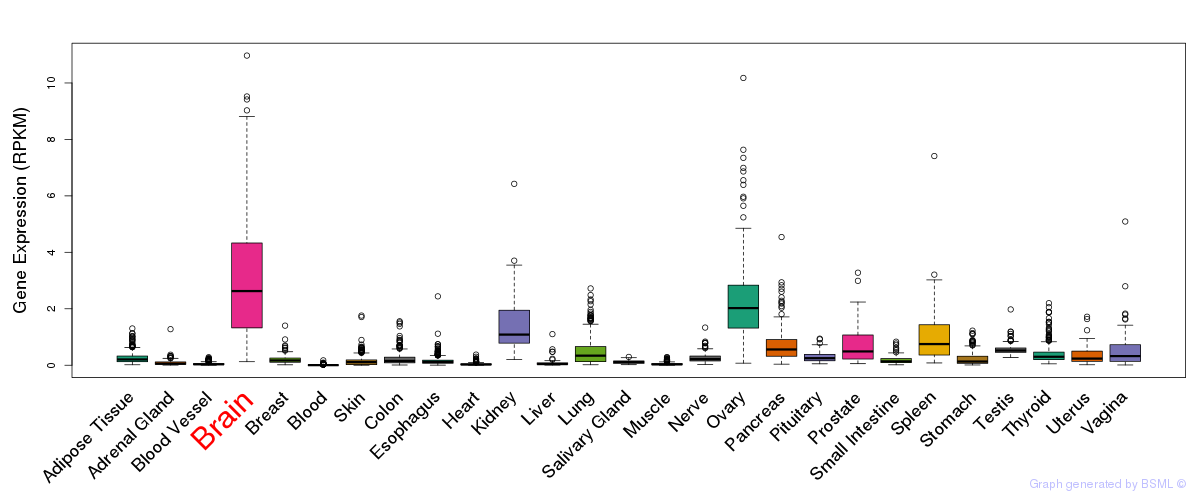
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BEGUM TARGETS OF PAX3 FOXO1 FUSION DN | 45 | 34 | All SZGR 2.0 genes in this pathway |
| WHITE NEUROBLASTOMA WITH 1P36.3 DELETION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| OKAWA NEUROBLASTOMA 1P36 31 DELETION | 22 | 19 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS DN | 35 | 22 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |