Gene Page: MAPK8
Summary ?
| GeneID | 5599 |
| Symbol | MAPK8 |
| Synonyms | JNK|JNK-46|JNK1|JNK1A2|JNK21B1/2|PRKM8|SAPK1|SAPK1c |
| Description | mitogen-activated protein kinase 8 |
| Reference | MIM:601158|HGNC:HGNC:6881|Ensembl:ENSG00000107643|HPRD:03100|Vega:OTTHUMG00000018172 |
| Gene type | protein-coding |
| Map location | 10q11.22 |
| Pascal p-value | 0.206 |
| TADA p-value | 0.009 |
| Fetal beta | 0.955 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0312 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MAPK8 | chr10 | 49628258 | G | A | NM_002750 NM_139046 NM_139047 NM_139049 | p.171G>S p.171G>S p.171G>S p.171G>S | missense missense missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
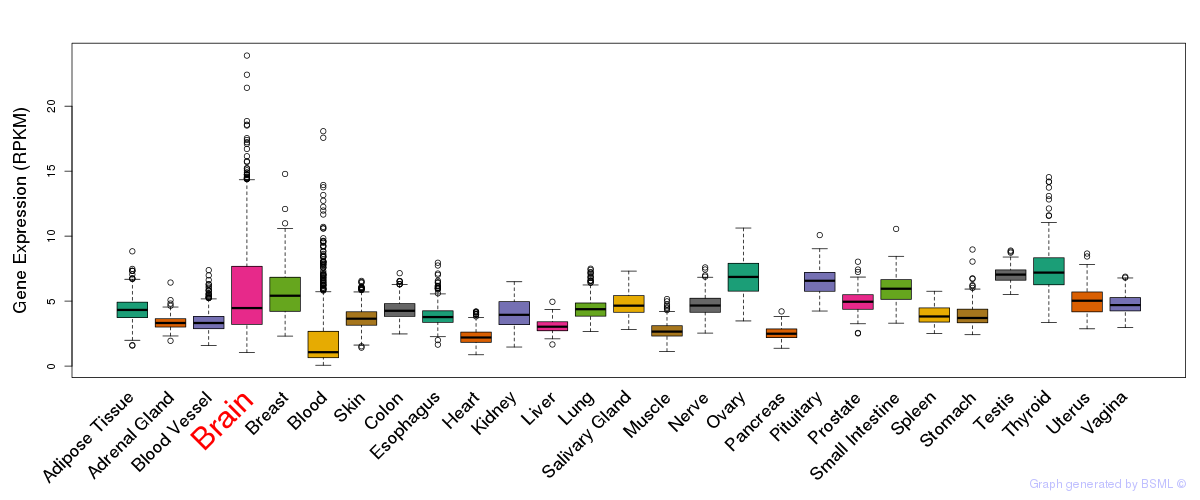
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LDHA | 0.87 | 0.88 |
| PPP2CA | 0.86 | 0.84 |
| PAIP2 | 0.85 | 0.85 |
| C20orf3 | 0.85 | 0.81 |
| GRSF1 | 0.85 | 0.84 |
| UBAP1 | 0.84 | 0.82 |
| KLHL12 | 0.84 | 0.80 |
| SHQ1 | 0.84 | 0.83 |
| MTO1 | 0.84 | 0.84 |
| PSMD7 | 0.84 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.65 | -0.49 |
| AF347015.2 | -0.63 | -0.50 |
| AF347015.18 | -0.63 | -0.56 |
| MT-CO2 | -0.62 | -0.50 |
| AF347015.8 | -0.62 | -0.50 |
| AF347015.26 | -0.62 | -0.49 |
| AF347015.33 | -0.60 | -0.50 |
| MT-CYB | -0.59 | -0.48 |
| AF347015.31 | -0.59 | -0.48 |
| AF347015.15 | -0.58 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004707 | MAP kinase activity | IEA | - | |
| GO:0004705 | JUN kinase activity | IDA | 8654373 | |
| GO:0005515 | protein binding | IPI | 10490659 |11238452 |15334056 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 14967141 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007258 | JUN phosphorylation | IDA | 14967141 | |
| GO:0006950 | response to stress | TAS | 8137421 | |
| GO:0007165 | signal transduction | TAS | 8137421 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 14764673 | |
| GO:0009411 | response to UV | IDA | 14967141 | |
| GO:0006928 | cell motion | TAS | 10912793 | |
| GO:0043066 | negative regulation of apoptosis | IDA | 14967141 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 9162092 |12591950 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 7535770 |9207092 |9405416 |11279118 |
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | - | HPRD | 7737129 |7737130 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western | BioGRID | 11432831 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Co-purification | BioGRID | 15659383 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | CDKN2A (p16INK4a) interacts with MAPK8 (JNK1) | BIND | 16007099 |
| COPS2 | ALIEN | CSN2 | SGN2 | TRIP15 | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | - | HPRD | 10585392 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD,BioGRID | 11432831 |
| DUSP1 | CL100 | HVH1 | MKP-1 | MKP1 | PTPN10 | dual specificity phosphatase 1 | - | HPRD,BioGRID | 11278799 |
| DUSP10 | MKP-5 | MKP5 | dual specificity phosphatase 10 | - | HPRD,BioGRID | 10391943 |10597297 |
| DUSP10 | MKP-5 | MKP5 | dual specificity phosphatase 10 | An unspecified isoform of MAPK8 interacts with DUSP10. This interaction was modelled on a demonstrated interaction between MAPK8 from an unspecified species and human DUSP10. | BIND | 10391943 |
| DUSP16 | KIAA1700 | MGC129701 | MGC129702 | MKP-7 | MKP7 | dual specificity phosphatase 16 | - | HPRD,BioGRID | 11489891 |
| DUSP22 | JKAP | JSP1 | MKPX | VHX | dual specificity phosphatase 22 | - | HPRD | 12138158 |
| DUSP22 | JKAP | JSP1 | MKPX | VHX | dual specificity phosphatase 22 | Affinity Capture-Western Biochemical Activity | BioGRID | 11346645 |
| DUSP4 | HVH2 | MKP-2 | MKP2 | TYP | dual specificity phosphatase 4 | Reconstituted Complex | BioGRID | 11387337 |
| ELK1 | - | ELK1, member of ETS oncogene family | An unspecified isoform of JNK1 interacts with and phosphorylates Elk1. | BIND | 9235954 |
| ELK1 | - | ELK1, member of ETS oncogene family | Biochemical Activity | BioGRID | 8586671 |
| ELK1 | - | ELK1, member of ETS oncogene family | Elk-1 is phosphorylated by JNK-1. This interaction was modeled on a demonstrated interaction between human Elk-1 and JNK-1 from an unspecified species. | BIND | 9020136 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | Sap-1a is phosphorylated by stress-activated JNK-1. This interaction was modeled on a demonstrated interaction between human Sap-1a and JNK-1 from an unspecified species. | BIND | 9020136 |
| ETV1 | DKFZp781L0674 | ER81 | MGC104699 | MGC120533 | MGC120534 | ets variant 1 | Biochemical Activity | BioGRID | 11551945 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Co-purification | BioGRID | 15659383 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| GSTP1 | DFN7 | FAEES3 | GST3 | PI | glutathione S-transferase pi 1 | - | HPRD,BioGRID | 11279197 |12646564 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 8137421 |11525649 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | - | HPRD,BioGRID | 12058026 |
| IL27RA | CRL1 | IL27R | TCCR | WSX1 | zcytor1 | interleukin 27 receptor, alpha | - | HPRD | 12734330 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 10722755 |11606564 |12417588 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 8137421 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK1 phosphorylates c-Jun. This interaction was modeled on a demonstrated interaction between bovine JNK1 or JNK1 from an unspecified species, and c-Jun from an unspecified species. | BIND | 9374537 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK1beta2 interacts with c-Jun. | BIND | 12167088 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | c-Jun is phosphorylated by JNK-1. This interaction was modeled on a demonstrated interaction between c-Jun and JNK-1, both from an unspecified species. | BIND | 9020136 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK1beta1 interacts with c-Jun. | BIND | 12167088 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK1alpha1 interacts with c-Jun. | BIND | 12167088 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | MAPK8 (JNK1) phosphorylates JUN (c-Jun) | BIND | 16007099 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | JNK1alpha2 interacts with c-Jun. | BIND | 12167088 |
| JUNB | AP-1 | jun B proto-oncogene | - | HPRD,BioGRID | 9405416 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD,BioGRID | 12052834 |
| MAP1B | DKFZp686E1099 | DKFZp686F1345 | FLJ38954 | FUTSCH | MAP5 | microtubule-associated protein 1B | - | HPRD | 12689591 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD | 9661668 |11062067 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | MKK4 phosphorylates JNK1. This interaction was modeled on a demonstrated interaction between MKK4 from an unspecified species and JNK1 from an unpsecified species. | BIND | 12659851 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | Biochemical Activity Phenotypic Enhancement Reconstituted Complex | BioGRID | 9207092 |11279118 |11707464 |12391307 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | - | HPRD,BioGRID | 9207092 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | MKK7 phosphorylates JNK1. This interaction was modeled on a demonstrated interaction between MKK7 from an unspecified species and JNK1 from an unspecified species. | BIND | 12659851 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 9405400 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | - | HPRD,BioGRID | 10713157 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 11865055 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | - | HPRD,BioGRID | 9733513 |
| MAPK8IP2 | IB2 | JIP2 | PRKM8IPL | mitogen-activated protein kinase 8 interacting protein 2 | - | HPRD | 10756100 |
| MAPK8IP3 | DKFZp762N1113 | FLJ00027 | JIP3 | JSAP1 | KIAA1066 | SYD2 | mitogen-activated protein kinase 8 interacting protein 3 | - | HPRD,BioGRID | 10523642 |10629060 |12189133 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 10551811 |
| PAX2 | - | paired box 2 | - | HPRD,BioGRID | 11700324 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | Affinity Capture-Western | BioGRID | 11948398 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD,BioGRID | 11749722 |
| REL | C-Rel | v-rel reticuloendotheliosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 8621542 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| SH2D3A | NSP1 | SH2 domain containing 3A | Phenotypic Enhancement | BioGRID | 10187783 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | Sab interacts with JNK1-alpha-2. | BIND | 12167088 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | Sab interacts with JNK1-alpha-1. | BIND | 12167088 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | - | HPRD,BioGRID | 12167088 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | Sab interacts with JNK1-beta-2. | BIND | 12167088 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | Sab interacts with JNK1-beta-1. | BIND | 12167088 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD | 10601313 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | Affinity Capture-Western | BioGRID | 12121974 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | - | HPRD,BioGRID | 12121974 |
| SPAG9 | FLJ13450 | FLJ14006 | FLJ26141 | FLJ34602 | HLC4 | JLP | KIAA0516 | MGC117291 | MGC14967 | MGC74461 | PHET | PIG6 | sperm associated antigen 9 | - | HPRD,BioGRID | 12391307 |
| SPI1 | OF | PU.1 | SFPI1 | SPI-1 | SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | Biochemical Activity | BioGRID | 8632909 |
| SPIB | SPI-B | Spi-B transcription factor (Spi-1/PU.1 related) | - | HPRD,BioGRID | 8632909 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Co-purification | BioGRID | 15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
| TNFSF11 | CD254 | ODF | OPGL | OPTB2 | RANKL | TRANCE | hRANKL2 | sOdf | tumor necrosis factor (ligand) superfamily, member 11 | - | HPRD | 9396779 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 9393873|9732264 |
| WWOX | D16S432E | FOR | FRA16D | HHCMA56 | PRO0128 | WOX1 | WW domain containing oxidoreductase | - | HPRD | 12514174 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD | 11749722 |