Gene Page: PARD3
Summary ?
| GeneID | 56288 |
| Symbol | PARD3 |
| Synonyms | ASIP|Baz|PAR3|PAR3alpha|PARD-3|PARD3A|PPP1R118|SE2-5L16|SE2-5LT1|SE2-5T2 |
| Description | par-3 family cell polarity regulator |
| Reference | MIM:606745|HGNC:HGNC:16051|Ensembl:ENSG00000148498|HPRD:05994|Vega:OTTHUMG00000017948 |
| Gene type | protein-coding |
| Map location | 10p11.21 |
| Pascal p-value | 0.376 |
| Fetal beta | 0.167 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.991 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
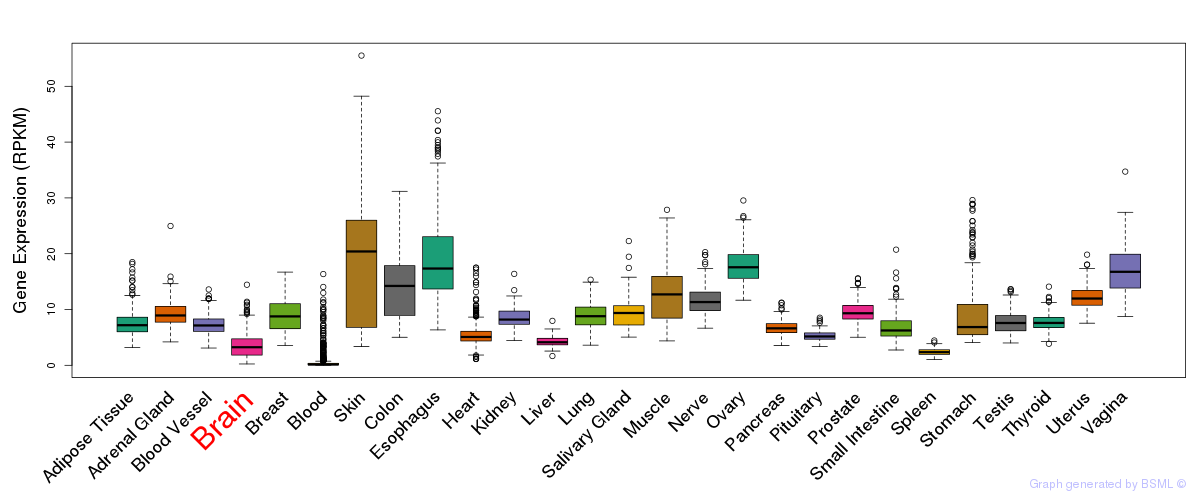
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | TAS | neuron, axon, neurite (GO term level: 12) | 14676191 |
| GO:0007205 | activation of protein kinase C activity | TAS | 11260256 | |
| GO:0006461 | protein complex assembly | TAS | 10934474 | |
| GO:0007163 | establishment or maintenance of cell polarity | TAS | 14676191 | |
| GO:0008356 | asymmetric cell division | TAS | 10934474 | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IDA | Brain (GO term level: 10) | 14676191 |
| GO:0005923 | tight junction | ISS | Brain (GO term level: 10) | - |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0005819 | spindle | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005938 | cell cortex | IEA | - | |
| GO:0005913 | cell-cell adherens junction | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | - | HPRD | 10934474 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| ECT2 | FLJ10461 | MGC138291 | epithelial cell transforming sequence 2 oncogene | Affinity Capture-Western | BioGRID | 15254234 |
| F11R | CD321 | JAM | JAM-1 | JAM-A | JAM1 | JAMA | JCAM | KAT | PAM-1 | F11 receptor | - | HPRD | 11447115 |
| F11R | CD321 | JAM | JAM-1 | JAM-A | JAM1 | JAMA | JCAM | KAT | PAM-1 | F11 receptor | Reconstituted Complex | BioGRID | 12953056 |
| JAM2 | C21orf43 | CD322 | JAM-B | JAMB | PRO245 | VE-JAM | VEJAM | junctional adhesion molecule 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12953056 |
| JAM3 | FLJ14529 | JAM-C | JAMC | junctional adhesion molecule 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12953056 |
| PARD6A | PAR-6A | PAR6 | PAR6C | PAR6alpha | TAX40 | TIP-40 | par-6 partitioning defective 6 homolog alpha (C. elegans) | Affinity Capture-Western | BioGRID | 12459187 |
| PARD6B | PAR6B | par-6 partitioning defective 6 homolog beta (C. elegans) | Affinity Capture-Western | BioGRID | 12459187 |
| PARD6G | FLJ45701 | PAR-6G | PAR6gamma | par-6 partitioning defective 6 homolog gamma (C. elegans) | Affinity Capture-Western | BioGRID | 12459187 |
| PLCB1 | FLJ45792 | PI-PLC | PLC-154 | PLC-I | PLC154 | phospholipase C, beta 1 (phosphoinositide-specific) | PLCB1 (PLC-beta-1) interacts with PARD3 (Par3). This interaction was modeled on a demonstrated interaction between rat PLCB1 and human Par3. | BIND | 15782111 |
| PLCB3 | FLJ37084 | phospholipase C, beta 3 (phosphatidylinositol-specific) | PLCB3 (PLC-beta-3) interacts with PARD3 (Par3). | BIND | 15782111 |
| PRKCI | DXS1179E | MGC26534 | PKCI | nPKC-iota | protein kinase C, iota | Affinity Capture-Western Reconstituted Complex | BioGRID | 12459187 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | Affinity Capture-Western | BioGRID | 12459187 |
| PVRL3 | CD113 | CDw113 | DKFZp566B0846 | FLJ90624 | PPR3 | PRR3 | PVRR3 | nectin-3 | poliovirus receptor-related 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12515806 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 12650946 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | - | HPRD | 12650946 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL JUNCTION ORGANIZATION | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME TIGHT JUNCTION INTERACTIONS | 29 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MCCOLLUM GELDANAMYCIN RESISTANCE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| WANG RECURRENT LIVER CANCER UP | 20 | 13 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |