Gene Page: PRPSAP1
Summary ?
| GeneID | 5635 |
| Symbol | PRPSAP1 |
| Synonyms | PAP39 |
| Description | phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| Reference | MIM:601249|HGNC:HGNC:9466|HPRD:03151| |
| Gene type | protein-coding |
| Map location | 17q24-q25 |
| Pascal p-value | 0.018 |
| Sherlock p-value | 0.756 |
| Fetal beta | 0.828 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14816748 | 17 | 74349897 | PRPSAP1 | 1.05E-7 | -0.008 | 2.3E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs346790 | chr17 | 74300294 | PRPSAP1 | 5635 | 0.02 | cis | ||
| rs447294 | chr17 | 74308124 | PRPSAP1 | 5635 | 0.06 | cis | ||
| rs421578 | chr17 | 74314772 | PRPSAP1 | 5635 | 0.01 | cis | ||
| rs164009 | 17 | 74283669 | PRPSAP1 | ENSG00000161542.12 | 1.679E-6 | 0.01 | 67341 | gtex_brain_ba24 |
| rs346785 | 17 | 74283769 | PRPSAP1 | ENSG00000161542.12 | 1.679E-6 | 0.01 | 67241 | gtex_brain_ba24 |
| rs4789272 | 17 | 74285783 | PRPSAP1 | ENSG00000161542.12 | 1.655E-6 | 0.01 | 65227 | gtex_brain_ba24 |
| rs17509527 | 17 | 74286173 | PRPSAP1 | ENSG00000161542.12 | 1.679E-6 | 0.01 | 64837 | gtex_brain_ba24 |
| rs164106 | 17 | 74290566 | PRPSAP1 | ENSG00000161542.12 | 1.677E-6 | 0.01 | 60444 | gtex_brain_ba24 |
| rs2945453 | 17 | 74291158 | PRPSAP1 | ENSG00000161542.12 | 9.946E-7 | 0.01 | 59852 | gtex_brain_ba24 |
| rs186985 | 17 | 74293388 | PRPSAP1 | ENSG00000161542.12 | 1.044E-6 | 0.01 | 57622 | gtex_brain_ba24 |
| rs4789276 | 17 | 74294598 | PRPSAP1 | ENSG00000161542.12 | 4.715E-7 | 0.01 | 56412 | gtex_brain_ba24 |
| rs12603578 | 17 | 74296079 | PRPSAP1 | ENSG00000161542.12 | 3.307E-7 | 0.01 | 54931 | gtex_brain_ba24 |
| rs2585747 | 17 | 74298257 | PRPSAP1 | ENSG00000161542.12 | 1.27E-7 | 0.01 | 52753 | gtex_brain_ba24 |
| rs443970 | 17 | 74303370 | PRPSAP1 | ENSG00000161542.12 | 1.198E-6 | 0.01 | 47640 | gtex_brain_ba24 |
| rs379503 | 17 | 74308358 | PRPSAP1 | ENSG00000161542.12 | 5.083E-7 | 0.01 | 42652 | gtex_brain_ba24 |
| rs2279057 | 17 | 74309425 | PRPSAP1 | ENSG00000161542.12 | 5.615E-7 | 0.01 | 41585 | gtex_brain_ba24 |
| rs2279056 | 17 | 74309474 | PRPSAP1 | ENSG00000161542.12 | 5.615E-7 | 0.01 | 41536 | gtex_brain_ba24 |
| rs407281 | 17 | 74311343 | PRPSAP1 | ENSG00000161542.12 | 5.942E-7 | 0.01 | 39667 | gtex_brain_ba24 |
| rs446662 | 17 | 74313542 | PRPSAP1 | ENSG00000161542.12 | 9.371E-7 | 0.01 | 37468 | gtex_brain_ba24 |
| rs186985 | 17 | 74293388 | PRPSAP1 | ENSG00000161542.12 | 2.192E-7 | 0 | 57622 | gtex_brain_putamen_basal |
| rs4789276 | 17 | 74294598 | PRPSAP1 | ENSG00000161542.12 | 3.63E-7 | 0 | 56412 | gtex_brain_putamen_basal |
| rs12603578 | 17 | 74296079 | PRPSAP1 | ENSG00000161542.12 | 2.011E-6 | 0 | 54931 | gtex_brain_putamen_basal |
| rs379503 | 17 | 74308358 | PRPSAP1 | ENSG00000161542.12 | 2.546E-7 | 0 | 42652 | gtex_brain_putamen_basal |
| rs2279057 | 17 | 74309425 | PRPSAP1 | ENSG00000161542.12 | 2.966E-7 | 0 | 41585 | gtex_brain_putamen_basal |
| rs2279056 | 17 | 74309474 | PRPSAP1 | ENSG00000161542.12 | 2.966E-7 | 0 | 41536 | gtex_brain_putamen_basal |
| rs407281 | 17 | 74311343 | PRPSAP1 | ENSG00000161542.12 | 2.414E-7 | 0 | 39667 | gtex_brain_putamen_basal |
| rs446662 | 17 | 74313542 | PRPSAP1 | ENSG00000161542.12 | 2.36E-6 | 0 | 37468 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
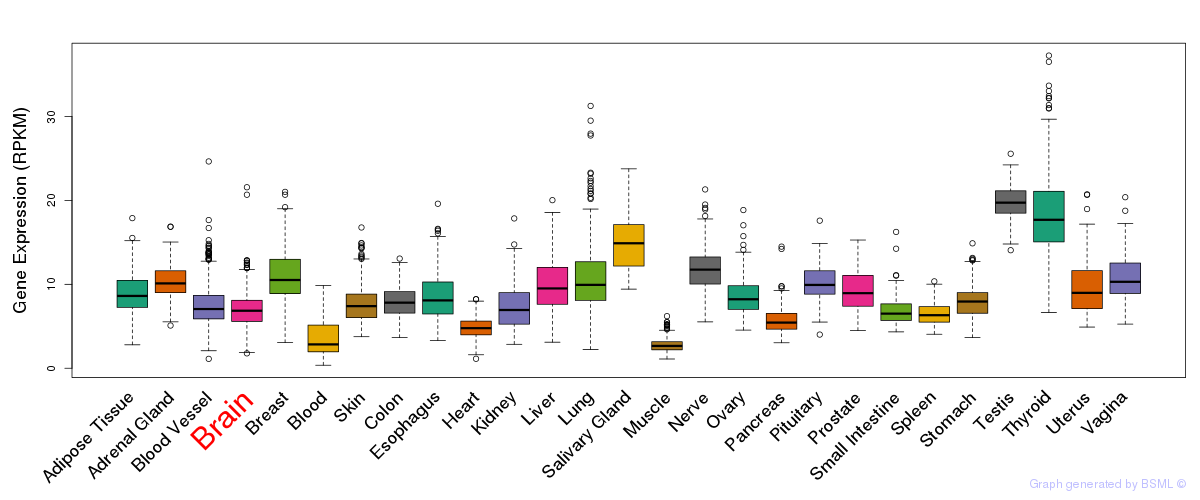
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD2BP2 | FWP010 | LIN1 | Snu40 | U5-52K | CD2 (cytoplasmic tail) binding protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Affinity Capture-MS | BioGRID | 17353931 |
| NUAK1 | ARK5 | KIAA0537 | NUAK family, SNF1-like kinase, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PLEKHF2 | FLJ13187 | PHAFIN2 | ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 | Two-hybrid | BioGRID | 16189514 |
| PRKRA | DYT16 | HSD14 | PACT | RAX | protein kinase, interferon-inducible double stranded RNA dependent activator | Affinity Capture-MS | BioGRID | 17353931 |
| PRPS1 | ARTS | CMTX5 | KIAA0967 | PPRibP | PRSI | phosphoribosyl pyrophosphate synthetase 1 | - | HPRD,BioGRID | 9366267 |
| PRPS2 | PRSII | phosphoribosyl pyrophosphate synthetase 2 | - | HPRD,BioGRID | 9366267 |
| PRPSAP1 | PAP39 | phosphoribosyl pyrophosphate synthetase-associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| PSPH | PSP | phosphoserine phosphatase | Affinity Capture-MS | BioGRID | 17353931 |
| PTP4A3 | PRL-3 | PRL-R | PRL3 | protein tyrosine phosphatase type IVA, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| TMBIM4 | CGI-119 | GAAP | S1R | ZPRO | transmembrane BAX inhibitor motif containing 4 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |