Gene Page: KLK6
Summary ?
| GeneID | 5653 |
| Symbol | KLK6 |
| Synonyms | Bssp|Klk7|PRSS18|PRSS9|SP59|hK6 |
| Description | kallikrein related peptidase 6 |
| Reference | MIM:602652|HGNC:HGNC:6367|Ensembl:ENSG00000167755|HPRD:04037|Vega:OTTHUMG00000183098 |
| Gene type | protein-coding |
| Map location | 19q13.3 |
| Pascal p-value | 0.068 |
| Fetal beta | -2.177 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6829546 | chr4 | 12587664 | KLK6 | 5653 | 0.19 | trans | ||
| rs6448932 | chr4 | 12595798 | KLK6 | 5653 | 0.11 | trans | ||
| rs7830259 | chr8 | 14087643 | KLK6 | 5653 | 0.02 | trans | ||
| rs8099473 | chr18 | 56200121 | KLK6 | 5653 | 0.05 | trans | ||
| rs8099488 | chr18 | 56200149 | KLK6 | 5653 | 0.17 | trans |
Section II. Transcriptome annotation
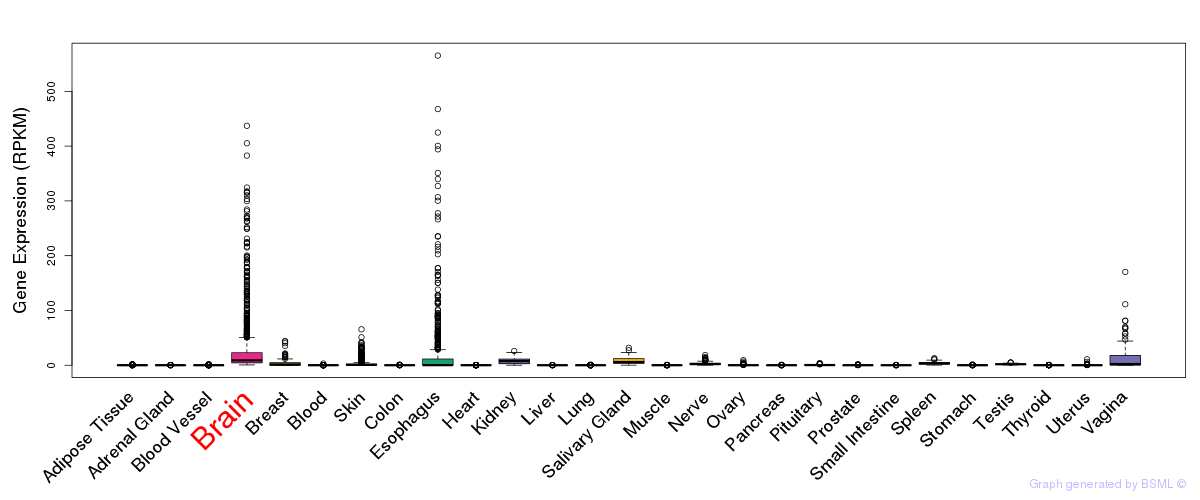
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMA3 | 0.91 | 0.89 |
| SARNP | 0.90 | 0.91 |
| C4orf27 | 0.90 | 0.90 |
| CWC15 | 0.89 | 0.89 |
| SSBP1 | 0.88 | 0.89 |
| SRP19 | 0.87 | 0.87 |
| UTP11L | 0.87 | 0.86 |
| SIP1 | 0.87 | 0.85 |
| LSM3 | 0.86 | 0.87 |
| EAPP | 0.86 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.69 | -0.68 |
| AF347015.15 | -0.68 | -0.66 |
| AF347015.26 | -0.68 | -0.68 |
| AF347015.33 | -0.66 | -0.64 |
| MT-CYB | -0.66 | -0.64 |
| AF347015.8 | -0.66 | -0.62 |
| MT-CO2 | -0.65 | -0.60 |
| AF347015.27 | -0.64 | -0.62 |
| FAM38A | -0.64 | -0.67 |
| AF347015.9 | -0.63 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004252 | serine-type endopeptidase activity | IEA | glutamate (GO term level: 7) | - |
| GO:0004252 | serine-type endopeptidase activity | NAS | glutamate (GO term level: 7) | 12878203 |
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 12709365 |12878203 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | NAS | Brain (GO term level: 6) | 12589961 |
| GO:0042552 | myelination | NAS | neuron, axon, Brain, oligodendrocyte (GO term level: 13) | 11668196 |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0009611 | response to wounding | NAS | 12016211 | |
| GO:0042982 | amyloid precursor protein metabolic process | NAS | 12709365 | |
| GO:0042246 | tissue regeneration | NAS | 12878203 | |
| GO:0016540 | protein autoprocessing | NAS | 12878203 | |
| GO:0042445 | hormone metabolic process | NAS | 11668196 | |
| GO:0030574 | collagen catabolic process | NAS | 12878203 | |
| GO:0045595 | regulation of cell differentiation | NAS | 11668196 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005792 | microsome | IEA | - | |
| GO:0005576 | extracellular region | IDA | 11668196 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 11668196 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP | 121 | 45 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| YAMASHITA METHYLATED IN PROSTATE CANCER | 57 | 29 | All SZGR 2.0 genes in this pathway |
| ENGELMANN CANCER PROGENITORS DN | 70 | 44 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG UP | 21 | 10 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 UP | 84 | 48 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |