Gene Page: DIABLO
Summary ?
| GeneID | 56616 |
| Symbol | DIABLO |
| Synonyms | DFNA64|SMAC |
| Description | diablo IAP-binding mitochondrial protein |
| Reference | MIM:605219|HGNC:HGNC:21528|Ensembl:ENSG00000184047|HPRD:05560|Vega:OTTHUMG00000157014 |
| Gene type | protein-coding |
| Map location | 12q24.31 |
| Sherlock p-value | 0.683 |
| Fetal beta | 0.338 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14894369 | 12 | 122709630 | DIABLO | 3.029E-4 | 0.248 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10738347 | chr9 | 13774022 | DIABLO | 56616 | 0.13 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
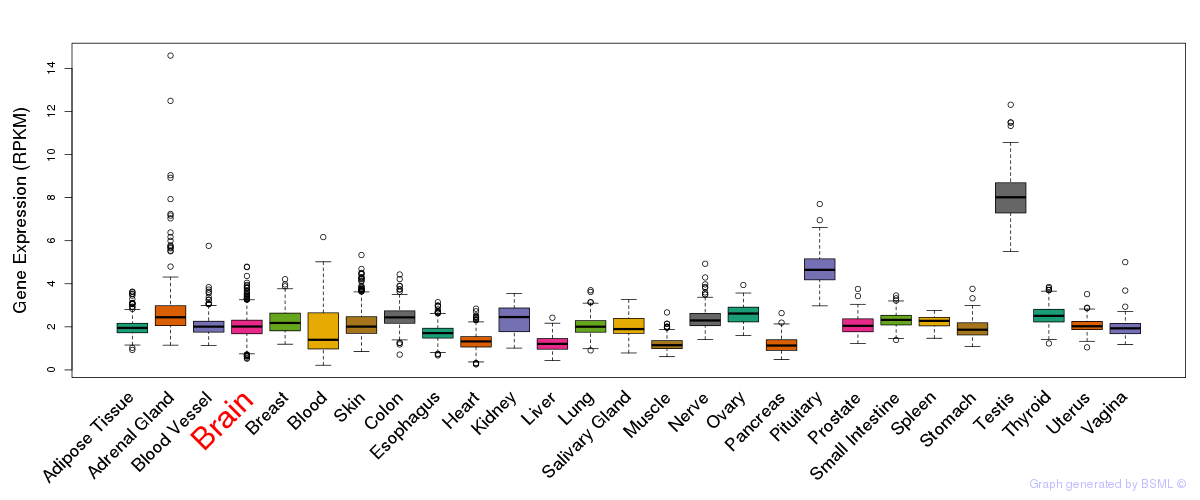
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | - | HPRD,BioGRID | 10929712 |
| BIRC3 | AIP1 | API2 | CIAP2 | HAIP1 | HIAP1 | MALT2 | MIHC | RNF49 | baculoviral IAP repeat-containing 3 | - | HPRD,BioGRID | 10929712 |
| BIRC5 | API4 | EPR-1 | baculoviral IAP repeat-containing 5 | - | HPRD,BioGRID | 12660240 |
| BIRC6 | APOLLON | BRUCE | FLJ13726 | FLJ13786 | KIAA1289 | baculoviral IAP repeat-containing 6 | Apollon interacts with and ubiquitinates SMAC. | BIND | 15300255 |
| BIRC6 | APOLLON | BRUCE | FLJ13726 | FLJ13786 | KIAA1289 | baculoviral IAP repeat-containing 6 | BRUCE interacts with SMAC. | BIND | 15200957 |
| DIABLO | DIABLO-S | FLJ10537 | FLJ25049 | SMAC | SMAC3 | diablo homolog (Drosophila) | - | HPRD,BioGRID | 10972280 |
| HTRA2 | OMI | PARK13 | PRSS25 | HtrA serine peptidase 2 | - | HPRD,BioGRID | 11604410 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | Smac interacts with LTBR. | BIND | 12571250 |
| LTBR | CD18 | D12S370 | LT-BETA-R | TNF-R-III | TNFCR | TNFR-RP | TNFR2-RP | TNFRSF3 | lymphotoxin beta receptor (TNFR superfamily, member 3) | - | HPRD,BioGRID | 12571250 |
| NAIP | BIRC1 | FLJ18088 | FLJ42520 | FLJ58811 | NLRB1 | psiNAIP | NLR family, apoptosis inhibitory protein | Affinity Capture-Western | BioGRID | 15280366 |
| NGFRAP1 | BEX3 | Bex | DXS6984E | HGR74 | NADE | nerve growth factor receptor (TNFRSF16) associated protein 1 | An unspecified isoform of NADE interacts with Smac. This interaction was modelled on a demonstrated interaction between mouse NADE and human Smac. | BIND | 15178455 |
| TNFSF14 | CD258 | HVEML | LIGHT | LTg | TR2 | tumor necrosis factor (ligand) superfamily, member 14 | Affinity Capture-MS | BioGRID | 12571250 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | Smac interacts with XIAP. | BIND | 15650747 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | XIAP interacts with and ubiquitinates SMAC. | BIND | 15300255 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | - | HPRD,BioGRID | 10929712 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA MITOCHONDRIA PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |