Gene Page: INPP5E
Summary ?
| GeneID | 56623 |
| Symbol | INPP5E |
| Synonyms | CORS1|CPD4|JBTS1|MORMS|PPI5PIV |
| Description | inositol polyphosphate-5-phosphatase E |
| Reference | MIM:613037|HGNC:HGNC:21474|Ensembl:ENSG00000148384|HPRD:17151|Vega:OTTHUMG00000020927 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.054 |
| Fetal beta | 0.675 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| INPP5E | chr9 | 139326418 | G | A | NM_019892 | . | silent | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15214069 | 9 | 139333582 | INPP5E | 1.14E-10 | -0.03 | 4.73E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12624267 | chr2 | 73986930 | INPP5E | 56623 | 0.19 | trans | ||
| rs4864466 | chr4 | 54403975 | INPP5E | 56623 | 0.16 | trans | ||
| rs3217906 | chr12 | 4405803 | INPP5E | 56623 | 0.13 | trans | ||
| rs10735662 | 9 | 139327869 | INPP5E | ENSG00000148384.11 | 1.191E-6 | 0.03 | 6405 | gtex_brain_ba24 |
| rs35735363 | 9 | 139330594 | INPP5E | ENSG00000148384.11 | 5.777E-7 | 0.03 | 3680 | gtex_brain_ba24 |
| rs1127152 | 9 | 139335599 | INPP5E | ENSG00000148384.11 | 1.146E-6 | 0.03 | -1325 | gtex_brain_ba24 |
| rs7874956 | 9 | 139336813 | INPP5E | ENSG00000148384.11 | 7.394E-7 | 0.03 | -2539 | gtex_brain_ba24 |
| rs36066734 | 9 | 139340351 | INPP5E | ENSG00000148384.11 | 5.966E-7 | 0.03 | -6077 | gtex_brain_ba24 |
| rs11145756 | 9 | 139364585 | INPP5E | ENSG00000148384.11 | 9.156E-7 | 0.03 | -30311 | gtex_brain_ba24 |
| rs6560632 | 9 | 139371786 | INPP5E | ENSG00000148384.11 | 5.775E-7 | 0.03 | -37512 | gtex_brain_ba24 |
| rs4378078 | 9 | 139375962 | INPP5E | ENSG00000148384.11 | 5.775E-7 | 0.03 | -41688 | gtex_brain_ba24 |
| rs7863766 | 9 | 139381314 | INPP5E | ENSG00000148384.11 | 5.75E-7 | 0.03 | -47040 | gtex_brain_ba24 |
| rs11145760 | 9 | 139382594 | INPP5E | ENSG00000148384.11 | 5.725E-7 | 0.03 | -48320 | gtex_brain_ba24 |
| rs11145912 | 9 | 139300876 | INPP5E | ENSG00000148384.11 | 7.755E-7 | 0 | 33398 | gtex_brain_putamen_basal |
| rs3087886 | 9 | 139301583 | INPP5E | ENSG00000148384.11 | 1.066E-7 | 0 | 32691 | gtex_brain_putamen_basal |
| rs11387728 | 9 | 139302777 | INPP5E | ENSG00000148384.11 | 5.119E-8 | 0 | 31497 | gtex_brain_putamen_basal |
| rs4487900 | 9 | 139303301 | INPP5E | ENSG00000148384.11 | 4.659E-9 | 0 | 30973 | gtex_brain_putamen_basal |
| rs10870146 | 9 | 139308054 | INPP5E | ENSG00000148384.11 | 3.274E-9 | 0 | 26220 | gtex_brain_putamen_basal |
| rs12380242 | 9 | 139310187 | INPP5E | ENSG00000148384.11 | 1.974E-8 | 0 | 24087 | gtex_brain_putamen_basal |
| rs3812584 | 9 | 139312719 | INPP5E | ENSG00000148384.11 | 1.246E-8 | 0 | 21555 | gtex_brain_putamen_basal |
| rs4604565 | 9 | 139313403 | INPP5E | ENSG00000148384.11 | 1.069E-7 | 0 | 20871 | gtex_brain_putamen_basal |
| rs10870164 | 9 | 139315682 | INPP5E | ENSG00000148384.11 | 3.687E-9 | 0 | 18592 | gtex_brain_putamen_basal |
| rs10870165 | 9 | 139316601 | INPP5E | ENSG00000148384.11 | 3.772E-7 | 0 | 17673 | gtex_brain_putamen_basal |
| rs7628 | 9 | 139318071 | INPP5E | ENSG00000148384.11 | 1.229E-7 | 0 | 16203 | gtex_brain_putamen_basal |
| rs10781538 | 9 | 139320929 | INPP5E | ENSG00000148384.11 | 1.063E-8 | 0 | 13345 | gtex_brain_putamen_basal |
| rs10781539 | 9 | 139320965 | INPP5E | ENSG00000148384.11 | 1.987E-6 | 0 | 13309 | gtex_brain_putamen_basal |
| rs10781540 | 9 | 139321836 | INPP5E | ENSG00000148384.11 | 7.791E-9 | 0 | 12438 | gtex_brain_putamen_basal |
| rs4259518 | 9 | 139322503 | INPP5E | ENSG00000148384.11 | 2.104E-9 | 0 | 11771 | gtex_brain_putamen_basal |
| rs8413 | 9 | 139323311 | INPP5E | ENSG00000148384.11 | 1.91E-9 | 0 | 10963 | gtex_brain_putamen_basal |
| rs1128874 | 9 | 139323424 | INPP5E | ENSG00000148384.11 | 7.989E-9 | 0 | 10850 | gtex_brain_putamen_basal |
| rs35763810 | 9 | 139323799 | INPP5E | ENSG00000148384.11 | 7.989E-9 | 0 | 10475 | gtex_brain_putamen_basal |
| rs68002978 | 9 | 139325641 | INPP5E | ENSG00000148384.11 | 1.02E-8 | 0 | 8633 | gtex_brain_putamen_basal |
| rs10870194 | 9 | 139327034 | INPP5E | ENSG00000148384.11 | 7.989E-9 | 0 | 7240 | gtex_brain_putamen_basal |
| rs10781542 | 9 | 139327439 | INPP5E | ENSG00000148384.11 | 7.989E-9 | 0 | 6835 | gtex_brain_putamen_basal |
| rs10735662 | 9 | 139327869 | INPP5E | ENSG00000148384.11 | 7.884E-8 | 0 | 6405 | gtex_brain_putamen_basal |
| rs10781543 | 9 | 139328369 | INPP5E | ENSG00000148384.11 | 6.813E-8 | 0 | 5905 | gtex_brain_putamen_basal |
| rs7851507 | 9 | 139328722 | INPP5E | ENSG00000148384.11 | 1.482E-6 | 0 | 5552 | gtex_brain_putamen_basal |
| rs7874956 | 9 | 139336813 | INPP5E | ENSG00000148384.11 | 4.374E-7 | 0 | -2539 | gtex_brain_putamen_basal |
| rs36066734 | 9 | 139340351 | INPP5E | ENSG00000148384.11 | 4.245E-7 | 0 | -6077 | gtex_brain_putamen_basal |
| rs10870065 | 9 | 139352946 | INPP5E | ENSG00000148384.11 | 7.982E-8 | 0 | -18672 | gtex_brain_putamen_basal |
| rs11145756 | 9 | 139364585 | INPP5E | ENSG00000148384.11 | 2.939E-8 | 0 | -30311 | gtex_brain_putamen_basal |
| rs6560632 | 9 | 139371786 | INPP5E | ENSG00000148384.11 | 3.52E-8 | 0 | -37512 | gtex_brain_putamen_basal |
| rs4378078 | 9 | 139375962 | INPP5E | ENSG00000148384.11 | 3.52E-8 | 0 | -41688 | gtex_brain_putamen_basal |
| rs4379550 | 9 | 139376426 | INPP5E | ENSG00000148384.11 | 1.543E-7 | 0 | -42152 | gtex_brain_putamen_basal |
| rs7045859 | 9 | 139377938 | INPP5E | ENSG00000148384.11 | 5.092E-7 | 0 | -43664 | gtex_brain_putamen_basal |
| rs7863766 | 9 | 139381314 | INPP5E | ENSG00000148384.11 | 3.363E-8 | 0 | -47040 | gtex_brain_putamen_basal |
| rs10870071 | 9 | 139382184 | INPP5E | ENSG00000148384.11 | 1.556E-7 | 0 | -47910 | gtex_brain_putamen_basal |
| rs11145760 | 9 | 139382594 | INPP5E | ENSG00000148384.11 | 3.345E-8 | 0 | -48320 | gtex_brain_putamen_basal |
| rs66481324 | 9 | 139383579 | INPP5E | ENSG00000148384.11 | 1.948E-7 | 0 | -49305 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
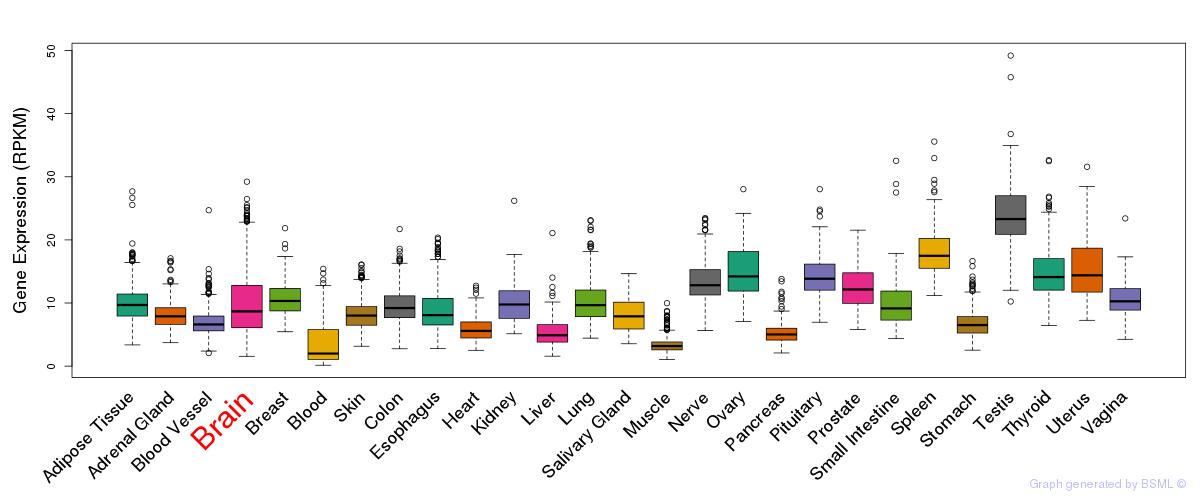
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |