Gene Page: NRIP3
Summary ?
| GeneID | 56675 |
| Symbol | NRIP3 |
| Synonyms | C11orf14|NY-SAR-105 |
| Description | nuclear receptor interacting protein 3 |
| Reference | MIM:613125|HGNC:HGNC:1167|Ensembl:ENSG00000175352|HPRD:14840|Vega:OTTHUMG00000165680 |
| Gene type | protein-coding |
| Map location | 11p15.3 |
| Pascal p-value | 0.447 |
| Sherlock p-value | 0.004 |
| Fetal beta | -2.347 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17014160 | chr1 | 206855799 | NRIP3 | 56675 | 0.02 | trans | ||
| snp_a-2090942 | 0 | NRIP3 | 56675 | 0.15 | trans | |||
| rs17014165 | chr1 | 206856485 | NRIP3 | 56675 | 0.15 | trans | ||
| rs17270544 | chr4 | 182529568 | NRIP3 | 56675 | 0.07 | trans | ||
| rs17211786 | chr4 | 182548238 | NRIP3 | 56675 | 0.16 | trans | ||
| rs2131824 | chr6 | 125775939 | NRIP3 | 56675 | 8.001E-5 | trans | ||
| rs2248 | chr6 | 125777720 | NRIP3 | 56675 | 8.001E-5 | trans | ||
| rs2249 | chr6 | 125777752 | NRIP3 | 56675 | 8.001E-5 | trans | ||
| rs1494142 | chr6 | 125778157 | NRIP3 | 56675 | 0 | trans | ||
| rs1494160 | chr6 | 125784791 | NRIP3 | 56675 | 8.001E-5 | trans | ||
| rs1494158 | chr6 | 125785156 | NRIP3 | 56675 | 8.001E-5 | trans | ||
| rs10115578 | chr9 | 3484372 | NRIP3 | 56675 | 0.01 | trans | ||
| rs2012359 | chr13 | 25078072 | NRIP3 | 56675 | 0.13 | trans | ||
| rs7887792 | chrX | 20508216 | NRIP3 | 56675 | 0.01 | trans |
Section II. Transcriptome annotation
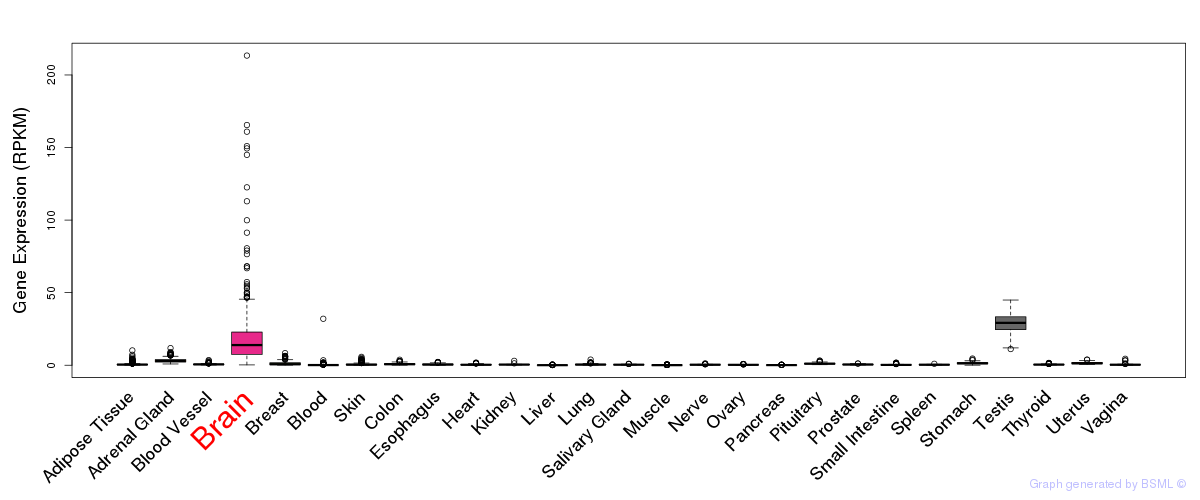
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN | 43 | 30 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR2 TARGETS UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL UP | 39 | 17 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION TOP20 UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |