Gene Page: EXOSC5
Summary ?
| GeneID | 56915 |
| Symbol | EXOSC5 |
| Synonyms | RRP41B|RRP46|Rrp46p|hRrp46p|p12B |
| Description | exosome component 5 |
| Reference | MIM:606492|HGNC:HGNC:24662|Ensembl:ENSG00000077348|HPRD:16222|Vega:OTTHUMG00000182759 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.104 |
| Sherlock p-value | 0.047 |
| Fetal beta | 0.052 |
| eGene | Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
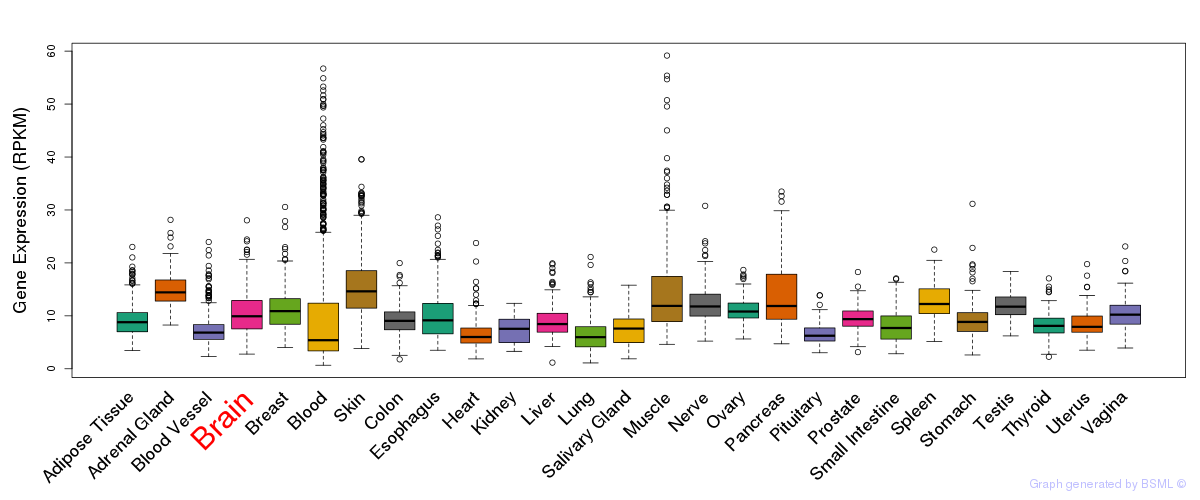
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EXOSC1 | CGI-108 | CSL4 | Csl4p | SKI4 | Ski4p | hCsl4p | p13 | exosome component 1 | hCsl4p interacts with hRrp46p. | BIND | 11812149 |
| EXOSC1 | CGI-108 | CSL4 | Csl4p | SKI4 | Ski4p | hCsl4p | p13 | exosome component 1 | - | HPRD,BioGRID | 11110791 |12419256|11812149 |12419256 |
| EXOSC1 | CGI-108 | CSL4 | Csl4p | SKI4 | Ski4p | hCsl4p | p13 | exosome component 1 | hRrp46p interacts with hCsl4p. | BIND | 12419256 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | - | HPRD | 15231747 |
| EXOSC3 | CGI-102 | MGC15120 | MGC723 | RP11-3J10.8 | RRP40 | Rrp40p | bA3J10.7 | hRrp40p | p10 | exosome component 3 | - | HPRD | 11110791 |12419256|12419256 |
| EXOSC3 | CGI-102 | MGC15120 | MGC723 | RP11-3J10.8 | RRP40 | Rrp40p | bA3J10.7 | hRrp40p | p10 | exosome component 3 | hRrp40p interacts with hRrp46p. | BIND | 12419256 |
| EXOSC8 | CIP3 | EAP2 | OIP2 | RP11-421P11.3 | RRP43 | Rrp43p | bA421P11.3 | p9 | exosome component 8 | hRrp46p interacts with OIP2. | BIND | 12419256 |
| EXOSC8 | CIP3 | EAP2 | OIP2 | RP11-421P11.3 | RRP43 | Rrp43p | bA421P11.3 | p9 | exosome component 8 | - | HPRD | 12419256 |
| KIAA1217 | DKFZp761L0424 | MGC31990 | SKT | KIAA1217 | Two-hybrid | BioGRID | 16189514 |
| LSM5 | FLJ12710 | YER146W | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| NUP210 | FLJ22389 | GP210 | KIAA0906 | POM210 | nucleoporin 210kDa | - | HPRD,BioGRID | 15231747 |
| PA2G4 | EBP1 | HG4-1 | p38-2G4 | proliferation-associated 2G4, 38kDa | - | HPRD | 15231747 |
| PKM2 | CTHBP | MGC3932 | OIP3 | PK3 | PKM | TCB | THBP1 | pyruvate kinase, muscle | - | HPRD,BioGRID | 15231747 |
| POLR2L | RBP10 | RPABC5 | RPB10 | RPB10beta | RPB7.6 | hRPB7.6 | hsRPB10b | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | - | HPRD | 15231747 |
| SFPQ | POMP100 | PSF | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | - | HPRD,BioGRID | 15231747 |
| ZNF558 | FLJ30932 | zinc finger protein 558 | - | HPRD | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME PERK REGULATED GENE EXPRESSION | 29 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF GENES BY ATF4 | 26 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY BRF1 | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY KSRP | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | 17 | 7 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |