Gene Page: NIT2
Summary ?
| GeneID | 56954 |
| Symbol | NIT2 |
| Synonyms | HEL-S-8a |
| Description | nitrilase family member 2 |
| Reference | MIM:616769|HGNC:HGNC:29878|HPRD:17634| |
| Gene type | protein-coding |
| Map location | 3q12.2 |
| Pascal p-value | 0.991 |
| Sherlock p-value | 0.599 |
| Fetal beta | -0.577 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.04047 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04359 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03429645 | 3 | 100053188 | NIT2 | 1.238E-4 | 0.588 | 0.03 | DMG:Wockner_2014 |
| cg24147849 | 3 | 100053372 | NIT2 | 1.03E-8 | -0.022 | 4.45E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9729667 | chr1 | 78349213 | NIT2 | 56954 | 0.1 | trans | ||
| rs16848937 | chr2 | 213482087 | NIT2 | 56954 | 0.2 | trans | ||
| rs1697139 | chr5 | 66511534 | NIT2 | 56954 | 0.15 | trans | ||
| rs10850170 | chr12 | 114031245 | NIT2 | 56954 | 0.19 | trans | ||
| rs7649598 | 3 | 100053083 | NIT2 | ENSG00000114021.7 | 4.882E-9 | 0 | -462 | gtex_brain_ba24 |
| rs11714045 | 3 | 100055125 | NIT2 | ENSG00000114021.7 | 1.162E-8 | 0 | 1580 | gtex_brain_ba24 |
| rs59560200 | 3 | 100059518 | NIT2 | ENSG00000114021.7 | 3.141E-8 | 0 | 5973 | gtex_brain_ba24 |
| rs35945684 | 3 | 100059666 | NIT2 | ENSG00000114021.7 | 2.31E-8 | 0 | 6121 | gtex_brain_ba24 |
| rs395399 | 3 | 100064005 | NIT2 | ENSG00000114021.7 | 2.663E-7 | 0 | 10460 | gtex_brain_ba24 |
| rs1045206 | 3 | 100064741 | NIT2 | ENSG00000114021.7 | 2.108E-7 | 0 | 11196 | gtex_brain_ba24 |
| rs2289505 | 3 | 100064829 | NIT2 | ENSG00000114021.7 | 1.162E-8 | 0 | 11284 | gtex_brain_ba24 |
| rs2289506 | 3 | 100064902 | NIT2 | ENSG00000114021.7 | 1.162E-8 | 0 | 11357 | gtex_brain_ba24 |
| rs1214375 | 3 | 100065632 | NIT2 | ENSG00000114021.7 | 2.086E-7 | 0 | 12087 | gtex_brain_ba24 |
| rs13082297 | 3 | 100071070 | NIT2 | ENSG00000114021.7 | 1.169E-8 | 0 | 17525 | gtex_brain_ba24 |
| rs62274146 | 3 | 100076980 | NIT2 | ENSG00000114021.7 | 1.016E-6 | 0 | 23435 | gtex_brain_ba24 |
| rs62274147 | 3 | 100091116 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 37571 | gtex_brain_ba24 |
| rs1486308 | 3 | 100093054 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 39509 | gtex_brain_ba24 |
| rs13071905 | 3 | 100094154 | NIT2 | ENSG00000114021.7 | 1.033E-6 | 0 | 40609 | gtex_brain_ba24 |
| rs13073252 | 3 | 100094208 | NIT2 | ENSG00000114021.7 | 1.033E-6 | 0 | 40663 | gtex_brain_ba24 |
| rs4928123 | 3 | 100094524 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 40979 | gtex_brain_ba24 |
| rs397788359 | 3 | 100100774 | NIT2 | ENSG00000114021.7 | 1.146E-6 | 0 | 47229 | gtex_brain_ba24 |
| rs3772693 | 3 | 100104114 | NIT2 | ENSG00000114021.7 | 1.112E-6 | 0 | 50569 | gtex_brain_ba24 |
| rs35281373 | 3 | 100111071 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 57526 | gtex_brain_ba24 |
| rs34645158 | 3 | 100111246 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 57701 | gtex_brain_ba24 |
| rs2289502 | 3 | 100121140 | NIT2 | ENSG00000114021.7 | 1.154E-6 | 0 | 67595 | gtex_brain_ba24 |
| rs4441690 | 3 | 100131897 | NIT2 | ENSG00000114021.7 | 2.079E-6 | 0 | 78352 | gtex_brain_ba24 |
| rs756103 | 3 | 100131976 | NIT2 | ENSG00000114021.7 | 2.102E-6 | 0 | 78431 | gtex_brain_ba24 |
| rs4928125 | 3 | 100141714 | NIT2 | ENSG00000114021.7 | 3.383E-6 | 0 | 88169 | gtex_brain_ba24 |
| rs11714045 | 3 | 100055125 | NIT2 | ENSG00000114021.7 | 6.423E-9 | 0 | 1580 | gtex_brain_putamen_basal |
| rs59560200 | 3 | 100059518 | NIT2 | ENSG00000114021.7 | 2.093E-9 | 0 | 5973 | gtex_brain_putamen_basal |
| rs35945684 | 3 | 100059666 | NIT2 | ENSG00000114021.7 | 2.246E-9 | 0 | 6121 | gtex_brain_putamen_basal |
| rs62274141 | 3 | 100060779 | NIT2 | ENSG00000114021.7 | 8.78E-7 | 0 | 7234 | gtex_brain_putamen_basal |
| rs2289505 | 3 | 100064829 | NIT2 | ENSG00000114021.7 | 6.423E-9 | 0 | 11284 | gtex_brain_putamen_basal |
| rs2289506 | 3 | 100064902 | NIT2 | ENSG00000114021.7 | 6.423E-9 | 0 | 11357 | gtex_brain_putamen_basal |
| rs13082297 | 3 | 100071070 | NIT2 | ENSG00000114021.7 | 6.423E-9 | 0 | 17525 | gtex_brain_putamen_basal |
| rs62274146 | 3 | 100076980 | NIT2 | ENSG00000114021.7 | 2.521E-8 | 0 | 23435 | gtex_brain_putamen_basal |
| rs62274147 | 3 | 100091116 | NIT2 | ENSG00000114021.7 | 5.822E-8 | 0 | 37571 | gtex_brain_putamen_basal |
| rs1486308 | 3 | 100093054 | NIT2 | ENSG00000114021.7 | 5.846E-8 | 0 | 39509 | gtex_brain_putamen_basal |
| rs13071905 | 3 | 100094154 | NIT2 | ENSG00000114021.7 | 5.854E-8 | 0 | 40609 | gtex_brain_putamen_basal |
| rs13073252 | 3 | 100094208 | NIT2 | ENSG00000114021.7 | 5.854E-8 | 0 | 40663 | gtex_brain_putamen_basal |
| rs4928123 | 3 | 100094524 | NIT2 | ENSG00000114021.7 | 5.854E-8 | 0 | 40979 | gtex_brain_putamen_basal |
| rs397788359 | 3 | 100100774 | NIT2 | ENSG00000114021.7 | 3.994E-8 | 0 | 47229 | gtex_brain_putamen_basal |
| rs3772693 | 3 | 100104114 | NIT2 | ENSG00000114021.7 | 3.838E-8 | 0 | 50569 | gtex_brain_putamen_basal |
| rs35281373 | 3 | 100111071 | NIT2 | ENSG00000114021.7 | 3.836E-8 | 0 | 57526 | gtex_brain_putamen_basal |
| rs34645158 | 3 | 100111246 | NIT2 | ENSG00000114021.7 | 3.836E-8 | 0 | 57701 | gtex_brain_putamen_basal |
| rs2289502 | 3 | 100121140 | NIT2 | ENSG00000114021.7 | 3.685E-8 | 0 | 67595 | gtex_brain_putamen_basal |
| rs4441690 | 3 | 100131897 | NIT2 | ENSG00000114021.7 | 3.437E-8 | 0 | 78352 | gtex_brain_putamen_basal |
| rs756103 | 3 | 100131976 | NIT2 | ENSG00000114021.7 | 3.454E-8 | 0 | 78431 | gtex_brain_putamen_basal |
| rs4928124 | 3 | 100136644 | NIT2 | ENSG00000114021.7 | 1.554E-7 | 0 | 83099 | gtex_brain_putamen_basal |
| rs4928125 | 3 | 100141714 | NIT2 | ENSG00000114021.7 | 3.227E-8 | 0 | 88169 | gtex_brain_putamen_basal |
| rs35925258 | 3 | 100173850 | NIT2 | ENSG00000114021.7 | 8.446E-7 | 0 | 120305 | gtex_brain_putamen_basal |
| rs55819532 | 3 | 100180901 | NIT2 | ENSG00000114021.7 | 1.097E-6 | 0 | 127356 | gtex_brain_putamen_basal |
| rs34064933 | 3 | 100183546 | NIT2 | ENSG00000114021.7 | 1.011E-6 | 0 | 130001 | gtex_brain_putamen_basal |
| rs10936205 | 3 | 100187904 | NIT2 | ENSG00000114021.7 | 1.766E-6 | 0 | 134359 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
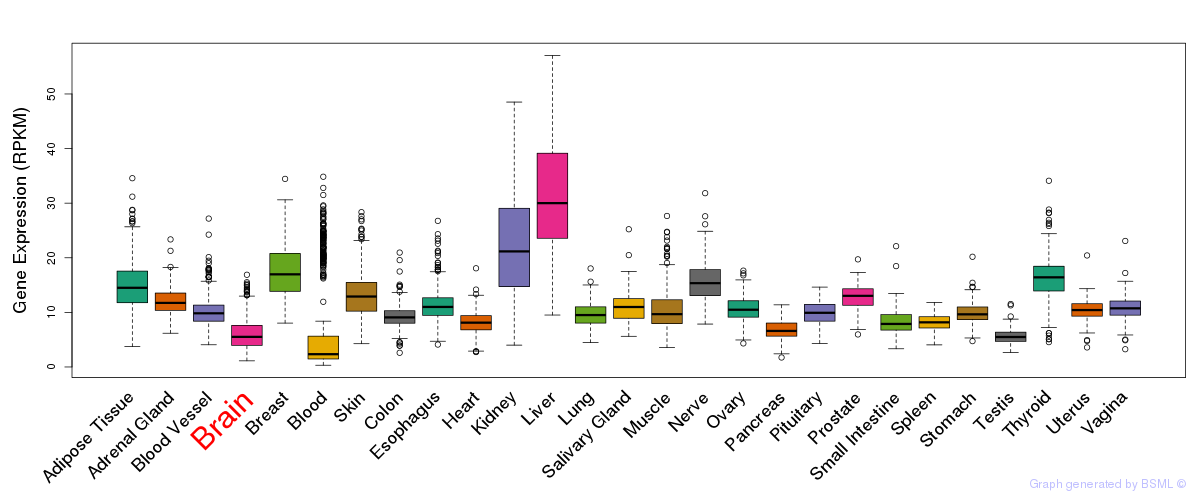
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| GO:0006807 | nitrogen compound metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALANINE ASPARTATE AND GLUTAMATE METABOLISM | 32 | 26 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |