Gene Page: PSMC3
Summary ?
| GeneID | 5702 |
| Symbol | PSMC3 |
| Synonyms | TBP1 |
| Description | proteasome 26S subunit, ATPase 3 |
| Reference | MIM:186852|HGNC:HGNC:9549|Ensembl:ENSG00000165916|HPRD:01733|Vega:OTTHUMG00000167692 |
| Gene type | protein-coding |
| Map location | 11p11.2 |
| Pascal p-value | 0.023 |
| Sherlock p-value | 0.595 |
| Fetal beta | -0.322 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.01 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08722104 | 11 | 47448306 | PSMC3 | 3.73E-10 | -0.019 | 7.47E-7 | DMG:Jaffe_2016 |
| cg23920472 | 11 | 47448172 | PSMC3 | 1.09E-8 | -0.011 | 4.61E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4752918 | 11 | 48253251 | PSMC3 | ENSG00000165916.4 | 3.201E-6 | 0.01 | -805258 | gtex_brain_ba24 |
| rs4500448 | 11 | 48253520 | PSMC3 | ENSG00000165916.4 | 2.606E-6 | 0.01 | -805527 | gtex_brain_ba24 |
| rs10838838 | 11 | 48253895 | PSMC3 | ENSG00000165916.4 | 2.576E-6 | 0.01 | -805902 | gtex_brain_ba24 |
| rs11039600 | 11 | 48254551 | PSMC3 | ENSG00000165916.4 | 4.048E-6 | 0.01 | -806558 | gtex_brain_ba24 |
| rs7395726 | 11 | 48256442 | PSMC3 | ENSG00000165916.4 | 3.323E-6 | 0.01 | -808449 | gtex_brain_ba24 |
| rs11039637 | 11 | 48306980 | PSMC3 | ENSG00000165916.4 | 4.127E-6 | 0.01 | -858987 | gtex_brain_ba24 |
| rs12221998 | 11 | 48308762 | PSMC3 | ENSG00000165916.4 | 3.827E-6 | 0.01 | -860769 | gtex_brain_ba24 |
| rs12221999 | 11 | 48308763 | PSMC3 | ENSG00000165916.4 | 3.827E-6 | 0.01 | -860770 | gtex_brain_ba24 |
| rs11530190 | 11 | 48309157 | PSMC3 | ENSG00000165916.4 | 3.017E-7 | 0.01 | -861164 | gtex_brain_ba24 |
| rs11530193 | 11 | 48309239 | PSMC3 | ENSG00000165916.4 | 6.288E-6 | 0.01 | -861246 | gtex_brain_ba24 |
| rs11039644 | 11 | 48314317 | PSMC3 | ENSG00000165916.4 | 4.669E-6 | 0.01 | -866324 | gtex_brain_ba24 |
Section II. Transcriptome annotation
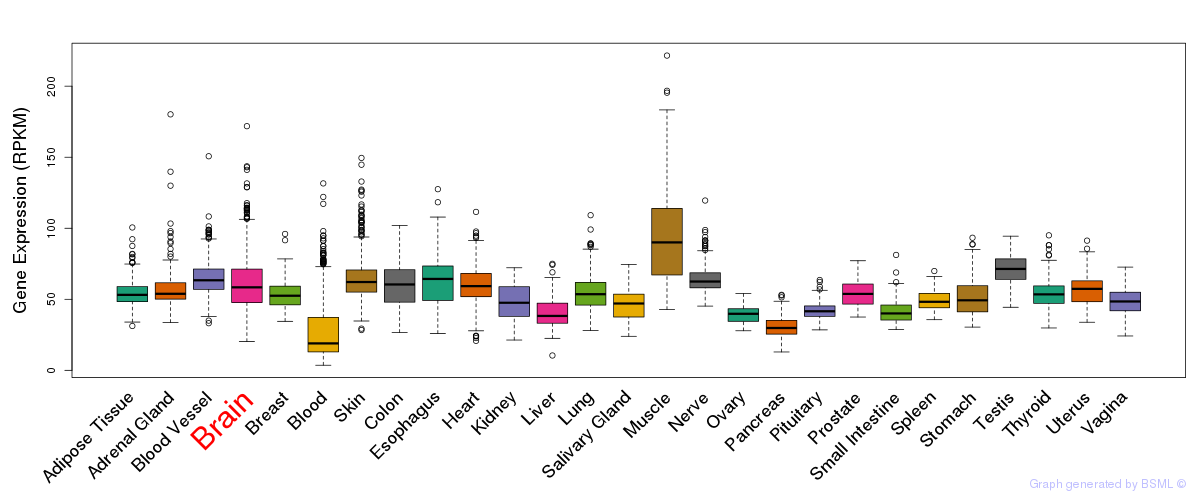
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNFAIP8L3 | 0.61 | 0.52 |
| WNT5A | 0.58 | 0.51 |
| NR3C2 | 0.57 | 0.61 |
| PKP2 | 0.56 | 0.51 |
| NEUROD1 | 0.54 | 0.32 |
| NRP1 | 0.54 | 0.46 |
| BNC1 | 0.53 | 0.21 |
| DUSP | 0.52 | 0.35 |
| MATN2 | 0.52 | 0.60 |
| C1QTNF3 | 0.52 | 0.24 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC098691.2 | -0.24 | -0.28 |
| AF347015.21 | -0.23 | -0.20 |
| MT-CO2 | -0.21 | -0.18 |
| PLA2G5 | -0.21 | -0.15 |
| AF347015.18 | -0.21 | -0.22 |
| AF347015.8 | -0.21 | -0.17 |
| CLEC2B | -0.19 | -0.17 |
| IL32 | -0.19 | -0.20 |
| MT-CYB | -0.19 | -0.15 |
| AF347015.33 | -0.19 | -0.13 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | TAS | 8419915 | |
| GO:0003714 | transcription corepressor activity | TAS | 2194290 | |
| GO:0005515 | protein binding | IPI | 17220478 |17353931 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0017111 | nucleoside-triphosphatase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001824 | blastocyst development | IEA | - | |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | EXP | 11285280 | |
| GO:0030163 | protein catabolic process | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 15029244 | |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 12791267 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000502 | proteasome complex | TAS | 8811196 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005634 | nucleus | TAS | 2194290 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | p14ARF interacts with TBP-1. | BIND | 14665636 |
| EPAS1 | ECYT4 | HIF2A | HLF | MOP2 | PASD2 | endothelial PAS domain protein 1 | Affinity Capture-Western | BioGRID | 14556007 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD,BioGRID | 14556007 |
| HTR1E | 5-HT1E | 5-hydroxytryptamine (serotonin) receptor 1E | Affinity Capture-MS | BioGRID | 17353931 |
| PAAF1 | FLJ11848 | PAAF | Rpn14 | WDR71 | proteasomal ATPase-associated factor 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC3 | MGC8487 | TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | - | HPRD | 11463857 |
| PSMC3IP | GT198 | HOP2 | HUMGT198A | TBPIP | PSMC3 interacting protein | - | HPRD | 9345291 |
| PSMC4 | MGC13687 | MGC23214 | MGC8570 | MIP224 | S6 | TBP7 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC4 | MGC13687 | MGC23214 | MGC8570 | MIP224 | S6 | TBP7 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | - | HPRD | 11463857 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | - | HPRD,BioGRID | 11463857 |
| PSMD10 | dJ889N15.2 | p28 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD13 | HSPC027 | Rpn9 | S11 | p40.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD6 | KIAA0107 | Rpn7 | S10 | SGA-113M | p44S10 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| STX11 | FHL4 | HLH4 | HPLH4 | syntaxin 11 | Two-hybrid | BioGRID | 16169070 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Affinity Capture-MS | BioGRID | 14743216 |
| TXNRD2 | SELZ | TR | TR-BETA | TR3 | TRXR2 | thioredoxin reductase 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11463857 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | - | HPRD,BioGRID | 14556007 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PROTEASOME | 48 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PROTEASOME PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | 51 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | 48 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | 51 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | 53 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | 72 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | 64 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | 51 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | 56 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | 52 | 30 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING DN | 35 | 24 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| WHITEHURST PACLITAXEL SENSITIVITY | 39 | 19 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION DN | 49 | 38 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WONG PROTEASOME GENE MODULE | 49 | 35 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |