Gene Page: DDX24
Summary ?
| GeneID | 57062 |
| Symbol | DDX24 |
| Synonyms | - |
| Description | DEAD-box helicase 24 |
| Reference | MIM:606181|HGNC:HGNC:13266|Ensembl:ENSG00000089737|HPRD:05860|Vega:OTTHUMG00000171301 |
| Gene type | protein-coding |
| Map location | 14q32 |
| Pascal p-value | 0.032 |
| Sherlock p-value | 0.25 |
| Fetal beta | -0.596 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02181506 | 14 | 94856984 | DDX24 | 1.96E-8 | 6.403 | DMG:vanEijk_2014 | |
| cg24621042 | 14 | 94857275 | DDX24 | 5.57E-5 | 4.973 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
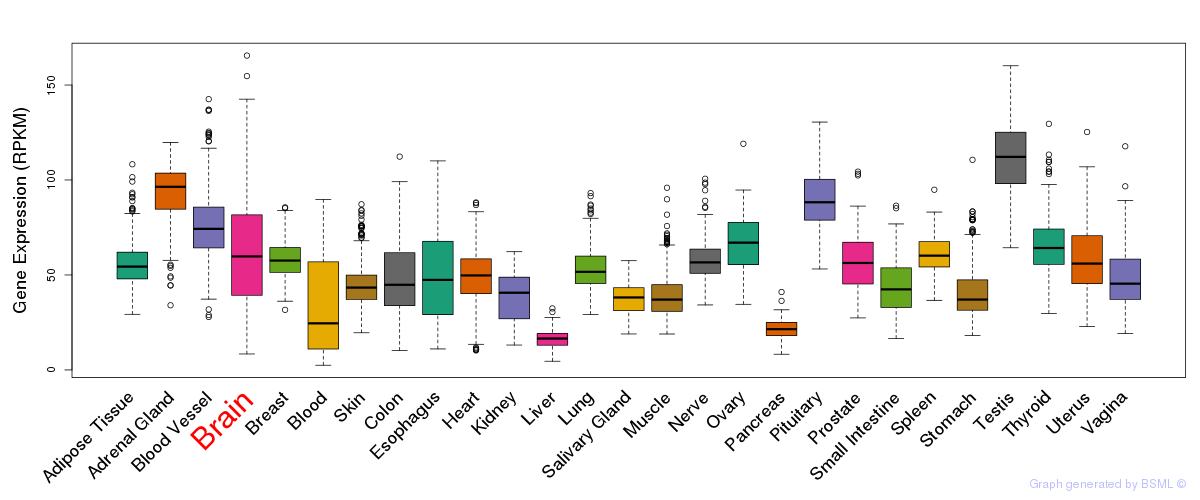
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCS1L | BCS | BCS1 | BJS | FLNMS | GRACILE | Hs.6719 | PTD | h-BCS | BCS1-like (yeast) | Two-hybrid | BioGRID | 16169070 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | Two-hybrid | BioGRID | 16169070 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | Two-hybrid | BioGRID | 16169070 |
| HSP90AB1 | D6S182 | FLJ26984 | HSP90-BETA | HSP90B | HSPC2 | HSPCB | heat shock protein 90kDa alpha (cytosolic), class B member 1 | Two-hybrid | BioGRID | 16169070 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | Two-hybrid | BioGRID | 16169070 |
| ITGB3BP | CENP-R | CENPR | HSU37139 | NRIF3 | TAP20 | integrin beta 3 binding protein (beta3-endonexin) | Two-hybrid | BioGRID | 16169070 |
| MKS1 | BBS13 | FLJ20345 | MES | MKS | Meckel syndrome, type 1 | Two-hybrid | BioGRID | 16169070 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 17353931 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Two-hybrid | BioGRID | 16169070 |
| POLA2 | FLJ21662 | FLJ37250 | polymerase (DNA directed), alpha 2 (70kD subunit) | Two-hybrid | BioGRID | 16169070 |
| SPTAN1 | (ALPHA)II-SPECTRIN | FLJ44613 | NEAS | spectrin, alpha, non-erythrocytic 1 (alpha-fodrin) | Two-hybrid | BioGRID | 16169070 |
| TPI1 | MGC88108 | TPI | triosephosphate isomerase 1 | Two-hybrid | BioGRID | 16169070 |
| TRDMT1 | DNMT2 | M.HsaIIP | PuMet | RNMT1 | tRNA aspartic acid methyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| VDAC3 | HD-VDAC3 | voltage-dependent anion channel 3 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |