Gene Page: RTN4
Summary ?
| GeneID | 57142 |
| Symbol | RTN4 |
| Synonyms | ASY|NI220/250|NOGO|NOGO-A|NOGOC|NSP|NSP-CL|Nbla00271|Nbla10545|Nogo-B|Nogo-C|RTN-X|RTN4-A|RTN4-B1|RTN4-B2|RTN4-C |
| Description | reticulon 4 |
| Reference | MIM:604475|HGNC:HGNC:14085|Ensembl:ENSG00000115310|HPRD:07259|Vega:OTTHUMG00000129337 |
| Gene type | protein-coding |
| Map location | 2p16.3 |
| Pascal p-value | 0.014 |
| Sherlock p-value | 0.606 |
| Fetal beta | -5.336 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26161045 | 2 | 55277080 | RTN4 | 1.634E-4 | -0.238 | 0.033 | DMG:Wockner_2014 |
| cg01555122 | 2 | 55276516 | RTN4 | 3.67E-9 | -0.026 | 2.35E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | RTN4 | 57142 | 0.17 | trans | ||
| rs11125553 | 2 | 55186928 | RTN4 | ENSG00000115310.13 | 1.777E-6 | 0 | 152829 | gtex_brain_ba24 |
| rs2580764 | 2 | 55196956 | RTN4 | ENSG00000115310.13 | 1.235E-6 | 0 | 142801 | gtex_brain_ba24 |
| rs2588510 | 2 | 55199248 | RTN4 | ENSG00000115310.13 | 1.149E-8 | 0 | 140509 | gtex_brain_ba24 |
| rs5831331 | 2 | 55199557 | RTN4 | ENSG00000115310.13 | 1.077E-8 | 0 | 140200 | gtex_brain_ba24 |
| rs2255112 | 2 | 55200800 | RTN4 | ENSG00000115310.13 | 1.149E-8 | 0 | 138957 | gtex_brain_ba24 |
| rs2588521 | 2 | 55203931 | RTN4 | ENSG00000115310.13 | 1.149E-8 | 0 | 135826 | gtex_brain_ba24 |
| rs2580765 | 2 | 55204028 | RTN4 | ENSG00000115310.13 | 1.149E-8 | 0 | 135729 | gtex_brain_ba24 |
| rs2588514 | 2 | 55209177 | RTN4 | ENSG00000115310.13 | 8.756E-8 | 0 | 130580 | gtex_brain_ba24 |
| rs2919129 | 2 | 55216545 | RTN4 | ENSG00000115310.13 | 1.735E-7 | 0 | 123212 | gtex_brain_ba24 |
| rs2015919 | 2 | 55218026 | RTN4 | ENSG00000115310.13 | 4.898E-7 | 0 | 121731 | gtex_brain_ba24 |
| rs2580772 | 2 | 55218805 | RTN4 | ENSG00000115310.13 | 3.034E-8 | 0 | 120952 | gtex_brain_ba24 |
| rs2580773 | 2 | 55219989 | RTN4 | ENSG00000115310.13 | 1.012E-6 | 0 | 119768 | gtex_brain_ba24 |
| rs2580776 | 2 | 55222132 | RTN4 | ENSG00000115310.13 | 1.87E-7 | 0 | 117625 | gtex_brain_ba24 |
| rs10210840 | 2 | 55226009 | RTN4 | ENSG00000115310.13 | 1.018E-6 | 0 | 113748 | gtex_brain_ba24 |
| rs7340476 | 2 | 55227346 | RTN4 | ENSG00000115310.13 | 2.087E-7 | 0 | 112411 | gtex_brain_ba24 |
| rs13387751 | 2 | 55227375 | RTN4 | ENSG00000115310.13 | 2.092E-7 | 0 | 112382 | gtex_brain_ba24 |
| rs6545466 | 2 | 55228392 | RTN4 | ENSG00000115310.13 | 8.722E-7 | 0 | 111365 | gtex_brain_ba24 |
| rs6760429 | 2 | 55228499 | RTN4 | ENSG00000115310.13 | 2.227E-7 | 0 | 111258 | gtex_brain_ba24 |
| rs12713284 | 2 | 55228861 | RTN4 | ENSG00000115310.13 | 2.175E-7 | 0 | 110896 | gtex_brain_ba24 |
| rs7562292 | 2 | 55230219 | RTN4 | ENSG00000115310.13 | 1.603E-7 | 0 | 109538 | gtex_brain_ba24 |
| rs6726158 | 2 | 55239202 | RTN4 | ENSG00000115310.13 | 8.511E-7 | 0 | 100555 | gtex_brain_ba24 |
| rs6755468 | 2 | 55240026 | RTN4 | ENSG00000115310.13 | 1.862E-7 | 0 | 99731 | gtex_brain_ba24 |
| rs10174838 | 2 | 55241216 | RTN4 | ENSG00000115310.13 | 8.31E-8 | 0 | 98541 | gtex_brain_ba24 |
| rs7584430 | 2 | 55241686 | RTN4 | ENSG00000115310.13 | 1.692E-7 | 0 | 98071 | gtex_brain_ba24 |
| rs35631428 | 2 | 55244438 | RTN4 | ENSG00000115310.13 | 1.731E-6 | 0 | 95319 | gtex_brain_ba24 |
| rs12477711 | 2 | 55245177 | RTN4 | ENSG00000115310.13 | 2.045E-6 | 0 | 94580 | gtex_brain_ba24 |
| rs2044291 | 2 | 55247954 | RTN4 | ENSG00000115310.13 | 2.218E-7 | 0 | 91803 | gtex_brain_ba24 |
| rs11687609 | 2 | 55192280 | RTN4 | ENSG00000115310.13 | 3.626E-6 | 0 | 147477 | gtex_brain_putamen_basal |
| rs2580764 | 2 | 55196956 | RTN4 | ENSG00000115310.13 | 5.459E-7 | 0 | 142801 | gtex_brain_putamen_basal |
| rs2255393 | 2 | 55197882 | RTN4 | ENSG00000115310.13 | 6.598E-11 | 0 | 141875 | gtex_brain_putamen_basal |
| rs2289747 | 2 | 55198385 | RTN4 | ENSG00000115310.13 | 4.372E-8 | 0 | 141372 | gtex_brain_putamen_basal |
| rs2241959 | 2 | 55198399 | RTN4 | ENSG00000115310.13 | 4.227E-7 | 0 | 141358 | gtex_brain_putamen_basal |
| rs887 | 2 | 55199068 | RTN4 | ENSG00000115310.13 | 1.656E-8 | 0 | 140689 | gtex_brain_putamen_basal |
| rs2588510 | 2 | 55199248 | RTN4 | ENSG00000115310.13 | 2.626E-9 | 0 | 140509 | gtex_brain_putamen_basal |
| rs5831331 | 2 | 55199557 | RTN4 | ENSG00000115310.13 | 9.963E-10 | 0 | 140200 | gtex_brain_putamen_basal |
| rs2255112 | 2 | 55200800 | RTN4 | ENSG00000115310.13 | 2.626E-9 | 0 | 138957 | gtex_brain_putamen_basal |
| rs2588521 | 2 | 55203931 | RTN4 | ENSG00000115310.13 | 2.626E-9 | 0 | 135826 | gtex_brain_putamen_basal |
| rs2580765 | 2 | 55204028 | RTN4 | ENSG00000115310.13 | 2.626E-9 | 0 | 135729 | gtex_brain_putamen_basal |
| rs2588514 | 2 | 55209177 | RTN4 | ENSG00000115310.13 | 2.502E-7 | 0 | 130580 | gtex_brain_putamen_basal |
| rs17418675 | 2 | 55210605 | RTN4 | ENSG00000115310.13 | 1.874E-6 | 0 | 129152 | gtex_brain_putamen_basal |
| rs12464447 | 2 | 55212495 | RTN4 | ENSG00000115310.13 | 2.489E-6 | 0 | 127262 | gtex_brain_putamen_basal |
| rs2919129 | 2 | 55216545 | RTN4 | ENSG00000115310.13 | 7.281E-8 | 0 | 123212 | gtex_brain_putamen_basal |
| rs2015919 | 2 | 55218026 | RTN4 | ENSG00000115310.13 | 1.153E-7 | 0 | 121731 | gtex_brain_putamen_basal |
| rs10496036 | 2 | 55218791 | RTN4 | ENSG00000115310.13 | 3.343E-6 | 0 | 120966 | gtex_brain_putamen_basal |
| rs2580772 | 2 | 55218805 | RTN4 | ENSG00000115310.13 | 7.281E-8 | 0 | 120952 | gtex_brain_putamen_basal |
| rs2580773 | 2 | 55219989 | RTN4 | ENSG00000115310.13 | 1.721E-7 | 0 | 119768 | gtex_brain_putamen_basal |
| rs17046570 | 2 | 55220584 | RTN4 | ENSG00000115310.13 | 3.343E-6 | 0 | 119173 | gtex_brain_putamen_basal |
| rs2580776 | 2 | 55222132 | RTN4 | ENSG00000115310.13 | 8.517E-8 | 0 | 117625 | gtex_brain_putamen_basal |
| rs55659423 | 2 | 55222630 | RTN4 | ENSG00000115310.13 | 3.55E-6 | 0 | 117127 | gtex_brain_putamen_basal |
| rs55698303 | 2 | 55222680 | RTN4 | ENSG00000115310.13 | 3.337E-6 | 0 | 117077 | gtex_brain_putamen_basal |
| rs2588518 | 2 | 55223282 | RTN4 | ENSG00000115310.13 | 5.4E-8 | 0 | 116475 | gtex_brain_putamen_basal |
| rs72795431 | 2 | 55225303 | RTN4 | ENSG00000115310.13 | 2.648E-6 | 0 | 114454 | gtex_brain_putamen_basal |
| rs10210840 | 2 | 55226009 | RTN4 | ENSG00000115310.13 | 8.612E-7 | 0 | 113748 | gtex_brain_putamen_basal |
| rs7340476 | 2 | 55227346 | RTN4 | ENSG00000115310.13 | 7.466E-8 | 0 | 112411 | gtex_brain_putamen_basal |
| rs13387751 | 2 | 55227375 | RTN4 | ENSG00000115310.13 | 7.469E-8 | 0 | 112382 | gtex_brain_putamen_basal |
| rs6545466 | 2 | 55228392 | RTN4 | ENSG00000115310.13 | 1.058E-7 | 0 | 111365 | gtex_brain_putamen_basal |
| rs6760429 | 2 | 55228499 | RTN4 | ENSG00000115310.13 | 7.515E-8 | 0 | 111258 | gtex_brain_putamen_basal |
| rs12713284 | 2 | 55228861 | RTN4 | ENSG00000115310.13 | 7.677E-8 | 0 | 110896 | gtex_brain_putamen_basal |
| rs35826668 | 2 | 55229508 | RTN4 | ENSG00000115310.13 | 5.384E-7 | 0 | 110249 | gtex_brain_putamen_basal |
| rs7562292 | 2 | 55230219 | RTN4 | ENSG00000115310.13 | 7.354E-8 | 0 | 109538 | gtex_brain_putamen_basal |
| rs6726158 | 2 | 55239202 | RTN4 | ENSG00000115310.13 | 7.889E-7 | 0 | 100555 | gtex_brain_putamen_basal |
| rs3806570 | 2 | 55239410 | RTN4 | ENSG00000115310.13 | 1.015E-7 | 0 | 100347 | gtex_brain_putamen_basal |
| rs6755468 | 2 | 55240026 | RTN4 | ENSG00000115310.13 | 1.045E-7 | 0 | 99731 | gtex_brain_putamen_basal |
| rs10174838 | 2 | 55241216 | RTN4 | ENSG00000115310.13 | 9.382E-8 | 0 | 98541 | gtex_brain_putamen_basal |
| rs7584430 | 2 | 55241686 | RTN4 | ENSG00000115310.13 | 1.139E-8 | 0 | 98071 | gtex_brain_putamen_basal |
| rs35631428 | 2 | 55244438 | RTN4 | ENSG00000115310.13 | 9.794E-7 | 0 | 95319 | gtex_brain_putamen_basal |
| rs17046594 | 2 | 55244538 | RTN4 | ENSG00000115310.13 | 2.188E-6 | 0 | 95219 | gtex_brain_putamen_basal |
| rs12477711 | 2 | 55245177 | RTN4 | ENSG00000115310.13 | 3.339E-8 | 0 | 94580 | gtex_brain_putamen_basal |
| rs4672005 | 2 | 55246551 | RTN4 | ENSG00000115310.13 | 2.484E-7 | 0 | 93206 | gtex_brain_putamen_basal |
| rs12464070 | 2 | 55247477 | RTN4 | ENSG00000115310.13 | 1.33E-7 | 0 | 92280 | gtex_brain_putamen_basal |
| rs2044291 | 2 | 55247954 | RTN4 | ENSG00000115310.13 | 1.258E-7 | 0 | 91803 | gtex_brain_putamen_basal |
| rs2902649 | 2 | 55248357 | RTN4 | ENSG00000115310.13 | 1.342E-7 | 0 | 91400 | gtex_brain_putamen_basal |
| rs7570629 | 2 | 55255935 | RTN4 | ENSG00000115310.13 | 1.04E-6 | 0 | 83822 | gtex_brain_putamen_basal |
| rs2968786 | 2 | 55264859 | RTN4 | ENSG00000115310.13 | 2.075E-6 | 0 | 74898 | gtex_brain_putamen_basal |
| rs141145795 | 2 | 55265634 | RTN4 | ENSG00000115310.13 | 2.152E-6 | 0 | 74123 | gtex_brain_putamen_basal |
| rs2968787 | 2 | 55265785 | RTN4 | ENSG00000115310.13 | 1.757E-7 | 0 | 73972 | gtex_brain_putamen_basal |
| rs2972097 | 2 | 55267545 | RTN4 | ENSG00000115310.13 | 2.143E-6 | 0 | 72212 | gtex_brain_putamen_basal |
| rs2860453 | 2 | 55270485 | RTN4 | ENSG00000115310.13 | 2.113E-7 | 0 | 69272 | gtex_brain_putamen_basal |
| rs2920893 | 2 | 55270629 | RTN4 | ENSG00000115310.13 | 1.678E-7 | 0 | 69128 | gtex_brain_putamen_basal |
| rs2920900 | 2 | 55280083 | RTN4 | ENSG00000115310.13 | 1.184E-6 | 0 | 59674 | gtex_brain_putamen_basal |
| rs2968790 | 2 | 55280594 | RTN4 | ENSG00000115310.13 | 6.018E-8 | 0 | 59163 | gtex_brain_putamen_basal |
| rs1822616 | 2 | 55283878 | RTN4 | ENSG00000115310.13 | 6.838E-8 | 0 | 55879 | gtex_brain_putamen_basal |
| rs2968793 | 2 | 55285024 | RTN4 | ENSG00000115310.13 | 4.883E-8 | 0 | 54733 | gtex_brain_putamen_basal |
| rs13011689 | 2 | 55287079 | RTN4 | ENSG00000115310.13 | 1.085E-7 | 0 | 52678 | gtex_brain_putamen_basal |
| rs2968796 | 2 | 55287835 | RTN4 | ENSG00000115310.13 | 5.102E-8 | 0 | 51922 | gtex_brain_putamen_basal |
| rs2968795 | 2 | 55287989 | RTN4 | ENSG00000115310.13 | 1.083E-7 | 0 | 51768 | gtex_brain_putamen_basal |
| rs565402040 | 2 | 55289148 | RTN4 | ENSG00000115310.13 | 8.566E-7 | 0 | 50609 | gtex_brain_putamen_basal |
| rs2968794 | 2 | 55290651 | RTN4 | ENSG00000115310.13 | 3.313E-7 | 0 | 49106 | gtex_brain_putamen_basal |
| rs11674808 | 2 | 55291671 | RTN4 | ENSG00000115310.13 | 1.418E-7 | 0 | 48086 | gtex_brain_putamen_basal |
| rs2972075 | 2 | 55292322 | RTN4 | ENSG00000115310.13 | 1.328E-7 | 0 | 47435 | gtex_brain_putamen_basal |
| rs34557804 | 2 | 55294571 | RTN4 | ENSG00000115310.13 | 1.313E-7 | 0 | 45186 | gtex_brain_putamen_basal |
| rs2972072 | 2 | 55295370 | RTN4 | ENSG00000115310.13 | 5.686E-8 | 0 | 44387 | gtex_brain_putamen_basal |
| rs2920878 | 2 | 55296415 | RTN4 | ENSG00000115310.13 | 1.338E-7 | 0 | 43342 | gtex_brain_putamen_basal |
| rs2968827 | 2 | 55297629 | RTN4 | ENSG00000115310.13 | 1.797E-7 | 0 | 42128 | gtex_brain_putamen_basal |
| rs2972071 | 2 | 55300749 | RTN4 | ENSG00000115310.13 | 6.272E-8 | 0 | 39008 | gtex_brain_putamen_basal |
| rs2972070 | 2 | 55301691 | RTN4 | ENSG00000115310.13 | 1.697E-7 | 0 | 38066 | gtex_brain_putamen_basal |
| rs2968825 | 2 | 55303042 | RTN4 | ENSG00000115310.13 | 3.822E-7 | 0 | 36715 | gtex_brain_putamen_basal |
| rs2968822 | 2 | 55307811 | RTN4 | ENSG00000115310.13 | 2.619E-7 | 0 | 31946 | gtex_brain_putamen_basal |
| rs2968821 | 2 | 55307865 | RTN4 | ENSG00000115310.13 | 9.645E-9 | 0 | 31892 | gtex_brain_putamen_basal |
| rs2920891 | 2 | 55308037 | RTN4 | ENSG00000115310.13 | 1.035E-8 | 0 | 31720 | gtex_brain_putamen_basal |
| rs2920882 | 2 | 55312270 | RTN4 | ENSG00000115310.13 | 3.034E-6 | 0 | 27487 | gtex_brain_putamen_basal |
| rs2968798 | 2 | 55314611 | RTN4 | ENSG00000115310.13 | 2.905E-6 | 0 | 25146 | gtex_brain_putamen_basal |
| rs2920862 | 2 | 55316938 | RTN4 | ENSG00000115310.13 | 1.943E-6 | 0 | 22819 | gtex_brain_putamen_basal |
| rs2968805 | 2 | 55319460 | RTN4 | ENSG00000115310.13 | 3.187E-6 | 0 | 20297 | gtex_brain_putamen_basal |
| rs2968809 | 2 | 55323803 | RTN4 | ENSG00000115310.13 | 1.865E-6 | 0 | 15954 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
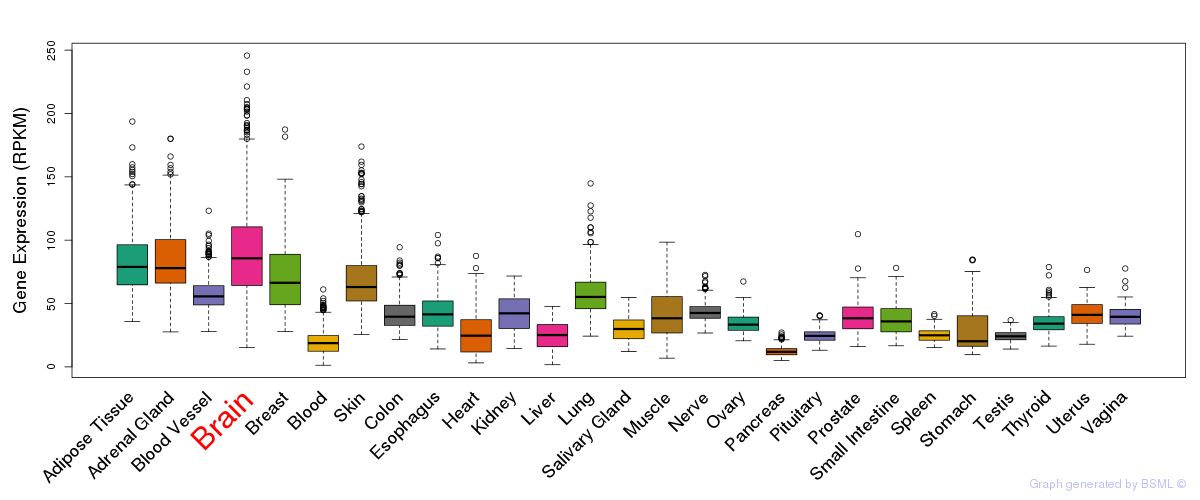
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11126360 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030517 | negative regulation of axon extension | IDA | axon (GO term level: 15) | 10667797 |
| GO:0019987 | negative regulation of anti-apoptosis | IMP | 11126360 | |
| GO:0042981 | regulation of apoptosis | NAS | 11126360 | |
| GO:0030334 | regulation of cell migration | IDA | 15034570 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005635 | nuclear envelope | IDA | 11126360 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005783 | endoplasmic reticulum | NAS | 11126360 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0030176 | integral to endoplasmic reticulum membrane | IDA | 10667797 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | RTN-XS interacts with Bcl-2. | BIND | 11126360 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 11126360 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD,BioGRID | 11126360 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | RTN-XS interacts with Bcl-XL. | BIND | 11126360 |
| CNTNAP1 | CASPR | CNTNAP | NRXN4 | P190 | contactin associated protein 1 | - | HPRD,BioGRID | 14592966 |
| COL4A3BP | CERT | CERTL | FLJ20597 | GPBP | STARD11 | collagen, type IV, alpha 3 (Goodpasture antigen) binding protein | Two-hybrid | BioGRID | 16189514 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | Two-hybrid | BioGRID | 16189514 |
| KCNA1 | AEMK | EA1 | HBK1 | HUK1 | KV1.1 | MBK1 | MGC126782 | MGC138385 | MK1 | RBK1 | potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | Affinity Capture-Western | BioGRID | 14592966 |
| KCNA2 | HBK5 | HK4 | HUKIV | KV1.2 | MGC50217 | MK2 | NGK1 | RBK2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | Affinity Capture-Western | BioGRID | 14592966 |
| LRCH4 | FLJ40101 | FLJ46315 | LRN | LRRN1 | LRRN4 | PP14183 | SAP25 | leucine-rich repeats and calponin homology (CH) domain containing 4 | Two-hybrid | BioGRID | 16189514 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Two-hybrid | BioGRID | 16169070 |
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | Two-hybrid | BioGRID | 16189514 |
| PLEKHF2 | FLJ13187 | PHAFIN2 | ZFYVE18 | pleckstrin homology domain containing, family F (with FYVE domain) member 2 | Two-hybrid | BioGRID | 16189514 |
| PRR13 | DKFZp564J157 | FLJ23818 | TXR1 | proline rich 13 | Two-hybrid | BioGRID | 16169070 |
| RAB33A | MGC1488 | RabS10 | RAB33A, member RAS oncogene family | Two-hybrid | BioGRID | 16189514 |
| RTN3 | ASYIP | HAP | NSPL2 | NSPLII | RTN3-A1 | reticulon 3 | Two-hybrid | BioGRID | 16189514 |
| RTN4IP1 | MGC12934 | NIMP | reticulon 4 interacting protein 1 | - | HPRD,BioGRID | 12067236 |
| RTN4IP1 | MGC12934 | NIMP | reticulon 4 interacting protein 1 | Nogo-A interacts with NIMP. | BIND | 12067236 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | - | HPRD | 11201742 |
| RTN4R | NGR | NOGOR | reticulon 4 receptor | Nogo-66 interacts with NgR. | BIND | 15694321 |
| SIRT2 | SIR2 | SIR2L | SIR2L2 | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SPG21 | ACP33 | BM-019 | GL010 | MASPARDIN | MAST | spastic paraplegia 21 (autosomal recessive, Mast syndrome) | Two-hybrid | BioGRID | 16189514 |
| UQCRC1 | D3S3191 | QCR1 | UQCR1 | ubiquinol-cytochrome c reductase core protein I | Nogo-A interacts with UQCRC1. | BIND | 12067236 |
| UQCRC1 | D3S3191 | QCR1 | UQCR1 | ubiquinol-cytochrome c reductase core protein I | - | HPRD,BioGRID | 12067236 |
| UQCRC2 | QCR2 | UQCR2 | ubiquinol-cytochrome c reductase core protein II | Nogo-A interacts with UQCRC2. | BIND | 12067236 |
| UQCRC2 | QCR2 | UQCR2 | ubiquinol-cytochrome c reductase core protein II | - | HPRD,BioGRID | 12067236 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG DN | 45 | 29 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 3 | 28 | 16 | All SZGR 2.0 genes in this pathway |
| LUI TARGETS OF PAX8 PPARG FUSION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| FUJIWARA PARK2 HEPATOCYTE PROLIFERATION DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| PETRETTO CARDIAC HYPERTROPHY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN | 85 | 56 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS DN | 30 | 15 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-132/212 | 509 | 515 | 1A | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-153 | 840 | 846 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 522 | 528 | m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-218 | 649 | 655 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-448 | 840 | 846 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-455 | 651 | 657 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-491 | 594 | 600 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.