Gene Page: PSMD13
Summary ?
| GeneID | 5719 |
| Symbol | PSMD13 |
| Synonyms | HSPC027|Rpn9|S11|p40.5 |
| Description | proteasome 26S subunit, non-ATPase 13 |
| Reference | MIM:603481|HGNC:HGNC:9558|Ensembl:ENSG00000185627|HPRD:04595|Vega:OTTHUMG00000119072 |
| Gene type | protein-coding |
| Map location | 11p15.5 |
| Pascal p-value | 0.967 |
| Sherlock p-value | 0.817 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.18:CC_BA10_disease_P=0.0054:HBB_BA9_fold_change=-1.21:HBB_BA9_disease_P=0.0116 |
| Fetal beta | 0.697 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7116130 | chr11 | 244128 | PSMD13 | 5719 | 0.12 | cis | ||
| rs1128322 | chr11 | 244196 | PSMD13 | 5719 | 0.05 | cis | ||
| rs532483 | chr11 | 257029 | PSMD13 | 5719 | 0.18 | cis | ||
| rs17156153 | chr11 | 270513 | PSMD13 | 5719 | 0.13 | cis | ||
| rs6751123 | chr2 | 62749568 | PSMD13 | 5719 | 0.04 | trans | ||
| rs56232710 | 11 | 202072 | PSMD13 | ENSG00000185627.13 | 3.208E-11 | 0 | -36926 | gtex_brain_ba24 |
| rs1045454 | 11 | 204228 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | -34770 | gtex_brain_ba24 |
| rs56357554 | 11 | 204715 | PSMD13 | ENSG00000185627.13 | 3.892E-11 | 0 | -34283 | gtex_brain_ba24 |
| rs3782120 | 11 | 206089 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | -32909 | gtex_brain_ba24 |
| rs7930823 | 11 | 206767 | PSMD13 | ENSG00000185627.13 | 2.294E-6 | 0 | -32231 | gtex_brain_ba24 |
| rs143657869 | 11 | 207254 | PSMD13 | ENSG00000185627.13 | 3.119E-11 | 0 | -31744 | gtex_brain_ba24 |
| rs6598075 | 11 | 207275 | PSMD13 | ENSG00000185627.13 | 3.479E-9 | 0 | -31723 | gtex_brain_ba24 |
| rs58692051 | 11 | 207410 | PSMD13 | ENSG00000185627.13 | 3.513E-9 | 0 | -31588 | gtex_brain_ba24 |
| rs201633036 | 11 | 209893 | PSMD13 | ENSG00000185627.13 | 2.231E-7 | 0 | -29105 | gtex_brain_ba24 |
| rs3832797 | 11 | 209894 | PSMD13 | ENSG00000185627.13 | 7.778E-8 | 0 | -29104 | gtex_brain_ba24 |
| rs200040945 | 11 | 209898 | PSMD13 | ENSG00000185627.13 | 4.115E-7 | 0 | -29100 | gtex_brain_ba24 |
| rs2293167 | 11 | 211644 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | -27354 | gtex_brain_ba24 |
| rs76552490 | 11 | 212015 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | -26983 | gtex_brain_ba24 |
| rs12574034 | 11 | 212262 | PSMD13 | ENSG00000185627.13 | 8.263E-12 | 0 | -26736 | gtex_brain_ba24 |
| rs1533825 | 11 | 214163 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | -24835 | gtex_brain_ba24 |
| rs1533824 | 11 | 214169 | PSMD13 | ENSG00000185627.13 | 7.618E-7 | 0 | -24829 | gtex_brain_ba24 |
| rs3825075 | 11 | 217140 | PSMD13 | ENSG00000185627.13 | 4.401E-12 | 0 | -21858 | gtex_brain_ba24 |
| rs511744 | 11 | 219089 | PSMD13 | ENSG00000185627.13 | 1.884E-12 | 0 | -19909 | gtex_brain_ba24 |
| rs6598074 | 11 | 219398 | PSMD13 | ENSG00000185627.13 | 8.346E-11 | 0 | -19600 | gtex_brain_ba24 |
| rs6598073 | 11 | 219452 | PSMD13 | ENSG00000185627.13 | 1.238E-6 | 0 | -19546 | gtex_brain_ba24 |
| rs6598072 | 11 | 219793 | PSMD13 | ENSG00000185627.13 | 1.552E-11 | 0 | -19205 | gtex_brain_ba24 |
| rs6598071 | 11 | 219800 | PSMD13 | ENSG00000185627.13 | 1.553E-11 | 0 | -19198 | gtex_brain_ba24 |
| rs10902107 | 11 | 220315 | PSMD13 | ENSG00000185627.13 | 2.102E-11 | 0 | -18683 | gtex_brain_ba24 |
| rs10794301 | 11 | 220401 | PSMD13 | ENSG00000185627.13 | 1.975E-11 | 0 | -18597 | gtex_brain_ba24 |
| rs6421986 | 11 | 221659 | PSMD13 | ENSG00000185627.13 | 5.346E-11 | 0 | -17339 | gtex_brain_ba24 |
| rs3782118 | 11 | 222620 | PSMD13 | ENSG00000185627.13 | 6.617E-9 | 0 | -16378 | gtex_brain_ba24 |
| rs3782116 | 11 | 223119 | PSMD13 | ENSG00000185627.13 | 2.573E-8 | 0 | -15879 | gtex_brain_ba24 |
| rs3782115 | 11 | 223272 | PSMD13 | ENSG00000185627.13 | 2.411E-7 | 0 | -15726 | gtex_brain_ba24 |
| rs7943246 | 11 | 225171 | PSMD13 | ENSG00000185627.13 | 6.465E-11 | 0 | -13827 | gtex_brain_ba24 |
| rs6598070 | 11 | 225466 | PSMD13 | ENSG00000185627.13 | 3.429E-11 | 0 | -13532 | gtex_brain_ba24 |
| rs7104764 | 11 | 229977 | PSMD13 | ENSG00000185627.13 | 1.452E-11 | 0 | -9021 | gtex_brain_ba24 |
| rs3829998 | 11 | 230751 | PSMD13 | ENSG00000185627.13 | 1.461E-11 | 0 | -8247 | gtex_brain_ba24 |
| rs10902109 | 11 | 230862 | PSMD13 | ENSG00000185627.13 | 1.463E-11 | 0 | -8136 | gtex_brain_ba24 |
| rs7483951 | 11 | 231758 | PSMD13 | ENSG00000185627.13 | 1.472E-11 | 0 | -7240 | gtex_brain_ba24 |
| rs10794302 | 11 | 234102 | PSMD13 | ENSG00000185627.13 | 8.147E-12 | 0 | -4896 | gtex_brain_ba24 |
| rs9795476 | 11 | 235894 | PSMD13 | ENSG00000185627.13 | 1.51E-11 | 0 | -3104 | gtex_brain_ba24 |
| rs11246029 | 11 | 236473 | PSMD13 | ENSG00000185627.13 | 1.243E-11 | 0 | -2525 | gtex_brain_ba24 |
| rs519592 | 11 | 236811 | PSMD13 | ENSG00000185627.13 | 8.51E-12 | 0 | -2187 | gtex_brain_ba24 |
| rs2272563 | 11 | 236871 | PSMD13 | ENSG00000185627.13 | 1.575E-11 | 0 | -2127 | gtex_brain_ba24 |
| rs1045288 | 11 | 237087 | PSMD13 | ENSG00000185627.13 | 2.058E-11 | 0 | -1911 | gtex_brain_ba24 |
| rs3817629 | 11 | 237312 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | -1686 | gtex_brain_ba24 |
| rs10902110 | 11 | 237648 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | -1350 | gtex_brain_ba24 |
| rs7481993 | 11 | 237875 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | -1123 | gtex_brain_ba24 |
| rs6598069 | 11 | 240664 | PSMD13 | ENSG00000185627.13 | 1.729E-11 | 0 | 1666 | gtex_brain_ba24 |
| rs7481518 | 11 | 240867 | PSMD13 | ENSG00000185627.13 | 6.176E-13 | 0 | 1869 | gtex_brain_ba24 |
| rs7482209 | 11 | 240868 | PSMD13 | ENSG00000185627.13 | 5.497E-13 | 0 | 1870 | gtex_brain_ba24 |
| rs7395328 | 11 | 242624 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 3626 | gtex_brain_ba24 |
| rs67547655 | 11 | 243017 | PSMD13 | ENSG00000185627.13 | 7.396E-13 | 0 | 4019 | gtex_brain_ba24 |
| rs59225278 | 11 | 243020 | PSMD13 | ENSG00000185627.13 | 7.537E-12 | 0 | 4022 | gtex_brain_ba24 |
| rs6598068 | 11 | 243092 | PSMD13 | ENSG00000185627.13 | 1.36E-11 | 0 | 4094 | gtex_brain_ba24 |
| rs6598067 | 11 | 243093 | PSMD13 | ENSG00000185627.13 | 1.084E-11 | 0 | 4095 | gtex_brain_ba24 |
| rs6598066 | 11 | 243185 | PSMD13 | ENSG00000185627.13 | 3.247E-11 | 0 | 4187 | gtex_brain_ba24 |
| rs6598065 | 11 | 243211 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4213 | gtex_brain_ba24 |
| rs6598064 | 11 | 243557 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4559 | gtex_brain_ba24 |
| rs6598063 | 11 | 243672 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4674 | gtex_brain_ba24 |
| rs6598062 | 11 | 243712 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4714 | gtex_brain_ba24 |
| rs6598061 | 11 | 243853 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4855 | gtex_brain_ba24 |
| rs6598060 | 11 | 243987 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 4989 | gtex_brain_ba24 |
| rs10902112 | 11 | 244106 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5108 | gtex_brain_ba24 |
| rs7107362 | 11 | 244108 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5110 | gtex_brain_ba24 |
| rs7128044 | 11 | 244115 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5117 | gtex_brain_ba24 |
| rs7116130 | 11 | 244129 | PSMD13 | ENSG00000185627.13 | 2.168E-11 | 0 | 5131 | gtex_brain_ba24 |
| rs7128029 | 11 | 244141 | PSMD13 | ENSG00000185627.13 | 2.031E-11 | 0 | 5143 | gtex_brain_ba24 |
| rs1128320 | 11 | 244167 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5169 | gtex_brain_ba24 |
| rs1128321 | 11 | 244171 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5173 | gtex_brain_ba24 |
| rs1128322 | 11 | 244197 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5199 | gtex_brain_ba24 |
| rs2272565 | 11 | 244414 | PSMD13 | ENSG00000185627.13 | 1.52E-11 | 0 | 5416 | gtex_brain_ba24 |
| rs398054895 | 11 | 244490 | PSMD13 | ENSG00000185627.13 | 2.425E-11 | 0 | 5492 | gtex_brain_ba24 |
| rs6598059 | 11 | 244961 | PSMD13 | ENSG00000185627.13 | 2.077E-10 | 0 | 5963 | gtex_brain_ba24 |
| rs7124022 | 11 | 244964 | PSMD13 | ENSG00000185627.13 | 2.094E-10 | 0 | 5966 | gtex_brain_ba24 |
| rs10902113 | 11 | 245523 | PSMD13 | ENSG00000185627.13 | 1.529E-11 | 0 | 6525 | gtex_brain_ba24 |
| rs78942883 | 11 | 245783 | PSMD13 | ENSG00000185627.13 | 1.651E-11 | 0 | 6785 | gtex_brain_ba24 |
| rs7482591 | 11 | 245861 | PSMD13 | ENSG00000185627.13 | 1.698E-11 | 0 | 6863 | gtex_brain_ba24 |
| rs10902114 | 11 | 246234 | PSMD13 | ENSG00000185627.13 | 1.666E-11 | 0 | 7236 | gtex_brain_ba24 |
| rs10794304 | 11 | 246456 | PSMD13 | ENSG00000185627.13 | 1.727E-11 | 0 | 7458 | gtex_brain_ba24 |
| rs10400248 | 11 | 247029 | PSMD13 | ENSG00000185627.13 | 8.087E-11 | 0 | 8031 | gtex_brain_ba24 |
| rs3830001 | 11 | 247200 | PSMD13 | ENSG00000185627.13 | 1.934E-6 | 0 | 8202 | gtex_brain_ba24 |
| rs56232710 | 11 | 202072 | PSMD13 | ENSG00000185627.13 | 1.934E-11 | 0 | -36926 | gtex_brain_putamen_basal |
| rs1045454 | 11 | 204228 | PSMD13 | ENSG00000185627.13 | 2.35E-11 | 0 | -34770 | gtex_brain_putamen_basal |
| rs56357554 | 11 | 204715 | PSMD13 | ENSG00000185627.13 | 2.87E-11 | 0 | -34283 | gtex_brain_putamen_basal |
| rs3782120 | 11 | 206089 | PSMD13 | ENSG00000185627.13 | 5.144E-12 | 0 | -32909 | gtex_brain_putamen_basal |
| rs7930823 | 11 | 206767 | PSMD13 | ENSG00000185627.13 | 3.313E-6 | 0 | -32231 | gtex_brain_putamen_basal |
| rs143657869 | 11 | 207254 | PSMD13 | ENSG00000185627.13 | 4.118E-11 | 0 | -31744 | gtex_brain_putamen_basal |
| rs6598075 | 11 | 207275 | PSMD13 | ENSG00000185627.13 | 2.347E-9 | 0 | -31723 | gtex_brain_putamen_basal |
| rs58692051 | 11 | 207410 | PSMD13 | ENSG00000185627.13 | 2.412E-9 | 0 | -31588 | gtex_brain_putamen_basal |
| rs201633036 | 11 | 209893 | PSMD13 | ENSG00000185627.13 | 9.837E-8 | 0 | -29105 | gtex_brain_putamen_basal |
| rs3832797 | 11 | 209894 | PSMD13 | ENSG00000185627.13 | 1.421E-8 | 0 | -29104 | gtex_brain_putamen_basal |
| rs200040945 | 11 | 209898 | PSMD13 | ENSG00000185627.13 | 6.291E-9 | 0 | -29100 | gtex_brain_putamen_basal |
| rs2293167 | 11 | 211644 | PSMD13 | ENSG00000185627.13 | 4.724E-12 | 0 | -27354 | gtex_brain_putamen_basal |
| rs76552490 | 11 | 212015 | PSMD13 | ENSG00000185627.13 | 4.724E-12 | 0 | -26983 | gtex_brain_putamen_basal |
| rs12574034 | 11 | 212262 | PSMD13 | ENSG00000185627.13 | 2.743E-12 | 0 | -26736 | gtex_brain_putamen_basal |
| rs1533825 | 11 | 214163 | PSMD13 | ENSG00000185627.13 | 4.724E-12 | 0 | -24835 | gtex_brain_putamen_basal |
| rs1533824 | 11 | 214169 | PSMD13 | ENSG00000185627.13 | 2.252E-11 | 0 | -24829 | gtex_brain_putamen_basal |
| rs3825075 | 11 | 217140 | PSMD13 | ENSG00000185627.13 | 6.304E-13 | 0 | -21858 | gtex_brain_putamen_basal |
| rs511744 | 11 | 219089 | PSMD13 | ENSG00000185627.13 | 5.842E-12 | 0 | -19909 | gtex_brain_putamen_basal |
| rs6598074 | 11 | 219398 | PSMD13 | ENSG00000185627.13 | 1.733E-11 | 0 | -19600 | gtex_brain_putamen_basal |
| rs6598073 | 11 | 219452 | PSMD13 | ENSG00000185627.13 | 8.114E-7 | 0 | -19546 | gtex_brain_putamen_basal |
| rs6598072 | 11 | 219793 | PSMD13 | ENSG00000185627.13 | 1.016E-12 | 0 | -19205 | gtex_brain_putamen_basal |
| rs6598071 | 11 | 219800 | PSMD13 | ENSG00000185627.13 | 1.015E-12 | 0 | -19198 | gtex_brain_putamen_basal |
| rs10902107 | 11 | 220315 | PSMD13 | ENSG00000185627.13 | 4.935E-12 | 0 | -18683 | gtex_brain_putamen_basal |
| rs10794301 | 11 | 220401 | PSMD13 | ENSG00000185627.13 | 2.677E-12 | 0 | -18597 | gtex_brain_putamen_basal |
| rs6421986 | 11 | 221659 | PSMD13 | ENSG00000185627.13 | 9.237E-12 | 0 | -17339 | gtex_brain_putamen_basal |
| rs3782118 | 11 | 222620 | PSMD13 | ENSG00000185627.13 | 1.097E-9 | 0 | -16378 | gtex_brain_putamen_basal |
| rs3782116 | 11 | 223119 | PSMD13 | ENSG00000185627.13 | 2.11E-8 | 0 | -15879 | gtex_brain_putamen_basal |
| rs3782115 | 11 | 223272 | PSMD13 | ENSG00000185627.13 | 5.41E-9 | 0 | -15726 | gtex_brain_putamen_basal |
| rs7943246 | 11 | 225171 | PSMD13 | ENSG00000185627.13 | 8.555E-12 | 0 | -13827 | gtex_brain_putamen_basal |
| rs6598070 | 11 | 225466 | PSMD13 | ENSG00000185627.13 | 1.541E-11 | 0 | -13532 | gtex_brain_putamen_basal |
| rs7104764 | 11 | 229977 | PSMD13 | ENSG00000185627.13 | 1.108E-12 | 0 | -9021 | gtex_brain_putamen_basal |
| rs3829998 | 11 | 230751 | PSMD13 | ENSG00000185627.13 | 1.11E-12 | 0 | -8247 | gtex_brain_putamen_basal |
| rs10902109 | 11 | 230862 | PSMD13 | ENSG00000185627.13 | 1.11E-12 | 0 | -8136 | gtex_brain_putamen_basal |
| rs7483951 | 11 | 231758 | PSMD13 | ENSG00000185627.13 | 1.112E-12 | 0 | -7240 | gtex_brain_putamen_basal |
| rs10794302 | 11 | 234102 | PSMD13 | ENSG00000185627.13 | 1.102E-12 | 0 | -4896 | gtex_brain_putamen_basal |
| rs9795476 | 11 | 235894 | PSMD13 | ENSG00000185627.13 | 1.117E-12 | 0 | -3104 | gtex_brain_putamen_basal |
| rs11246029 | 11 | 236473 | PSMD13 | ENSG00000185627.13 | 3.652E-12 | 0 | -2525 | gtex_brain_putamen_basal |
| rs519592 | 11 | 236811 | PSMD13 | ENSG00000185627.13 | 8.682E-11 | 0 | -2187 | gtex_brain_putamen_basal |
| rs2272563 | 11 | 236871 | PSMD13 | ENSG00000185627.13 | 1.124E-12 | 0 | -2127 | gtex_brain_putamen_basal |
| rs1045288 | 11 | 237087 | PSMD13 | ENSG00000185627.13 | 1.355E-12 | 0 | -1911 | gtex_brain_putamen_basal |
| rs3817629 | 11 | 237312 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | -1686 | gtex_brain_putamen_basal |
| rs10902110 | 11 | 237648 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | -1350 | gtex_brain_putamen_basal |
| rs7481993 | 11 | 237875 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | -1123 | gtex_brain_putamen_basal |
| rs6598069 | 11 | 240664 | PSMD13 | ENSG00000185627.13 | 1.11E-12 | 0 | 1666 | gtex_brain_putamen_basal |
| rs7481518 | 11 | 240867 | PSMD13 | ENSG00000185627.13 | 3.82E-14 | 0 | 1869 | gtex_brain_putamen_basal |
| rs7482209 | 11 | 240868 | PSMD13 | ENSG00000185627.13 | 3.688E-14 | 0 | 1870 | gtex_brain_putamen_basal |
| rs7395328 | 11 | 242624 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 3626 | gtex_brain_putamen_basal |
| rs67547655 | 11 | 243017 | PSMD13 | ENSG00000185627.13 | 1.842E-13 | 0 | 4019 | gtex_brain_putamen_basal |
| rs59225278 | 11 | 243020 | PSMD13 | ENSG00000185627.13 | 2.092E-11 | 0 | 4022 | gtex_brain_putamen_basal |
| rs6598068 | 11 | 243092 | PSMD13 | ENSG00000185627.13 | 4.601E-14 | 0 | 4094 | gtex_brain_putamen_basal |
| rs6598067 | 11 | 243093 | PSMD13 | ENSG00000185627.13 | 6.311E-14 | 0 | 4095 | gtex_brain_putamen_basal |
| rs6598066 | 11 | 243185 | PSMD13 | ENSG00000185627.13 | 1.716E-11 | 0 | 4187 | gtex_brain_putamen_basal |
| rs6598065 | 11 | 243211 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4213 | gtex_brain_putamen_basal |
| rs6598064 | 11 | 243557 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4559 | gtex_brain_putamen_basal |
| rs6598063 | 11 | 243672 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4674 | gtex_brain_putamen_basal |
| rs6598062 | 11 | 243712 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4714 | gtex_brain_putamen_basal |
| rs6598061 | 11 | 243853 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4855 | gtex_brain_putamen_basal |
| rs6598060 | 11 | 243987 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 4989 | gtex_brain_putamen_basal |
| rs10902112 | 11 | 244106 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5108 | gtex_brain_putamen_basal |
| rs7107362 | 11 | 244108 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5110 | gtex_brain_putamen_basal |
| rs7128044 | 11 | 244115 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5117 | gtex_brain_putamen_basal |
| rs7116130 | 11 | 244129 | PSMD13 | ENSG00000185627.13 | 1.546E-12 | 0 | 5131 | gtex_brain_putamen_basal |
| rs7128029 | 11 | 244141 | PSMD13 | ENSG00000185627.13 | 1.38E-12 | 0 | 5143 | gtex_brain_putamen_basal |
| rs1128320 | 11 | 244167 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5169 | gtex_brain_putamen_basal |
| rs1128321 | 11 | 244171 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5173 | gtex_brain_putamen_basal |
| rs1128322 | 11 | 244197 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5199 | gtex_brain_putamen_basal |
| rs2272565 | 11 | 244414 | PSMD13 | ENSG00000185627.13 | 1.118E-12 | 0 | 5416 | gtex_brain_putamen_basal |
| rs398054895 | 11 | 244490 | PSMD13 | ENSG00000185627.13 | 7.617E-13 | 0 | 5492 | gtex_brain_putamen_basal |
| rs2272566 | 11 | 244552 | PSMD13 | ENSG00000185627.13 | 1.516E-6 | 0 | 5554 | gtex_brain_putamen_basal |
| rs6598059 | 11 | 244961 | PSMD13 | ENSG00000185627.13 | 1.09E-11 | 0 | 5963 | gtex_brain_putamen_basal |
| rs7124022 | 11 | 244964 | PSMD13 | ENSG00000185627.13 | 6.743E-12 | 0 | 5966 | gtex_brain_putamen_basal |
| rs10902113 | 11 | 245523 | PSMD13 | ENSG00000185627.13 | 1.125E-12 | 0 | 6525 | gtex_brain_putamen_basal |
| rs78942883 | 11 | 245783 | PSMD13 | ENSG00000185627.13 | 1.305E-12 | 0 | 6785 | gtex_brain_putamen_basal |
| rs7482591 | 11 | 245861 | PSMD13 | ENSG00000185627.13 | 1.266E-12 | 0 | 6863 | gtex_brain_putamen_basal |
| rs10902114 | 11 | 246234 | PSMD13 | ENSG00000185627.13 | 1.363E-12 | 0 | 7236 | gtex_brain_putamen_basal |
| rs10794304 | 11 | 246456 | PSMD13 | ENSG00000185627.13 | 1.415E-12 | 0 | 7458 | gtex_brain_putamen_basal |
| rs10400248 | 11 | 247029 | PSMD13 | ENSG00000185627.13 | 2.492E-12 | 0 | 8031 | gtex_brain_putamen_basal |
| rs3830001 | 11 | 247200 | PSMD13 | ENSG00000185627.13 | 1.475E-6 | 0 | 8202 | gtex_brain_putamen_basal |
| rs6598056 | 11 | 252283 | PSMD13 | ENSG00000185627.13 | 1.373E-7 | 0 | 13285 | gtex_brain_putamen_basal |
| rs7930624 | 11 | 253841 | PSMD13 | ENSG00000185627.13 | 8.601E-7 | 0 | 14843 | gtex_brain_putamen_basal |
| rs6598054 | 11 | 254010 | PSMD13 | ENSG00000185627.13 | 8.601E-7 | 0 | 15012 | gtex_brain_putamen_basal |
| rs7121085 | 11 | 258523 | PSMD13 | ENSG00000185627.13 | 1.355E-6 | 0 | 19525 | gtex_brain_putamen_basal |
| rs7111364 | 11 | 258542 | PSMD13 | ENSG00000185627.13 | 1.355E-6 | 0 | 19544 | gtex_brain_putamen_basal |
| rs7950638 | 11 | 259695 | PSMD13 | ENSG00000185627.13 | 1.389E-6 | 0 | 20697 | gtex_brain_putamen_basal |
| rs367850937 | 11 | 273615 | PSMD13 | ENSG00000185627.13 | 1.32E-6 | 0 | 34617 | gtex_brain_putamen_basal |
| rs35409720 | 11 | 277365 | PSMD13 | ENSG00000185627.13 | 1.424E-6 | 0 | 38367 | gtex_brain_putamen_basal |
| rs200516324 | 11 | 277424 | PSMD13 | ENSG00000185627.13 | 2.73E-6 | 0 | 38426 | gtex_brain_putamen_basal |
| rs71487220 | 11 | 277425 | PSMD13 | ENSG00000185627.13 | 2.688E-6 | 0 | 38427 | gtex_brain_putamen_basal |
| rs79248624 | 11 | 278126 | PSMD13 | ENSG00000185627.13 | 6.879E-7 | 0 | 39128 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
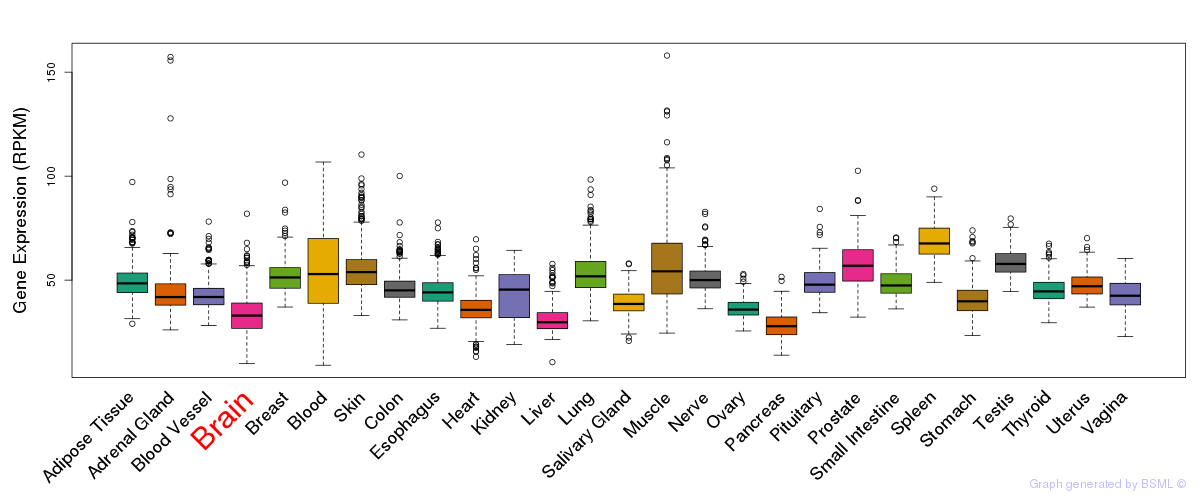
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CRB1 | LCA8 | RP12 | crumbs homolog 1 (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| FES | FPS | feline sarcoma oncogene | Affinity Capture-MS | BioGRID | 17353931 |
| MTHFD2 | NMDMC | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | Affinity Capture-MS | BioGRID | 17353931 |
| PSMA1 | HC2 | MGC14542 | MGC14575 | MGC14751 | MGC1667 | MGC21459 | MGC22853 | MGC23915 | NU | PROS30 | proteasome (prosome, macropain) subunit, alpha type, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMA3 | HC8 | MGC12306 | MGC32631 | PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMA6 | IOTA | MGC22756 | MGC2333 | MGC23846 | PROS27 | p27K | proteasome (prosome, macropain) subunit, alpha type, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMA7 | C6 | HSPC | MGC3755 | RC6-1 | XAPC7 | proteasome (prosome, macropain) subunit, alpha type, 7 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC1 | MGC24583 | MGC8541 | P26S4 | S4 | p56 | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC2 | MGC3004 | MSS1 | Nbla10058 | S7 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC3 | MGC8487 | TBP1 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC4 | MGC13687 | MGC23214 | MGC8570 | MIP224 | S6 | TBP7 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC5 | S8 | SUG1 | TBP10 | TRIP1 | p45 | p45/SUG | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMC6 | CADP44 | MGC12520 | P44 | SUG2 | p42 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD1 | MGC133040 | MGC133041 | P112 | Rpn2 | S1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD12 | MGC75406 | Rpn5 | p55 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD14 | PAD1 | POH1 | rpn11 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD2 | MGC14274 | P97 | Rpn1 | S2 | TRAP2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD3 | P58 | RPN3 | S3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD4 | AF | AF-1 | ASF | MCB1 | Rpn10 | S5A | pUB-R5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD6 | KIAA0107 | Rpn7 | S10 | SGA-113M | p44S10 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD7 | MOV34 | P40 | Rpn8 | S12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 | Affinity Capture-MS | BioGRID | 17353931 |
| PSMD8 | HIP6 | HYPF | MGC1660 | Nin1p | Rpn12 | S14 | p31 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | Affinity Capture-MS | BioGRID | 17353931 |
| SLC25A22 | FLJ13044 | GC1 | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | Affinity Capture-MS | BioGRID | 17353931 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Affinity Capture-MS | BioGRID | 14743216 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PROTEASOME | 48 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | 76 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ER PHAGOSOME PATHWAY | 61 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | 51 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | 48 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF APOPTOSIS | 58 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | 51 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | 53 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | 72 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | 64 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | 51 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | 56 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | 52 | 30 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 20 MCF10A | 14 | 9 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| SETLUR PROSTATE CANCER TMPRSS2 ERG FUSION UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |