Gene Page: ARID1B
Summary ?
| GeneID | 57492 |
| Symbol | ARID1B |
| Synonyms | 6A3-5|BAF250B|BRIGHT|DAN15|ELD/OSA1|MRD12|OSA2|P250R |
| Description | AT-rich interaction domain 1B |
| Reference | MIM:614556|HGNC:HGNC:18040|Ensembl:ENSG00000049618|HPRD:10660|Vega:OTTHUMG00000015890 |
| Gene type | protein-coding |
| Map location | 6q25.1 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.448 |
| Fetal beta | 0.746 |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARID1B | chr6 | 157495166 | T | C | NM_017519 NM_020732 | p.1004M>T p.1017M>T | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
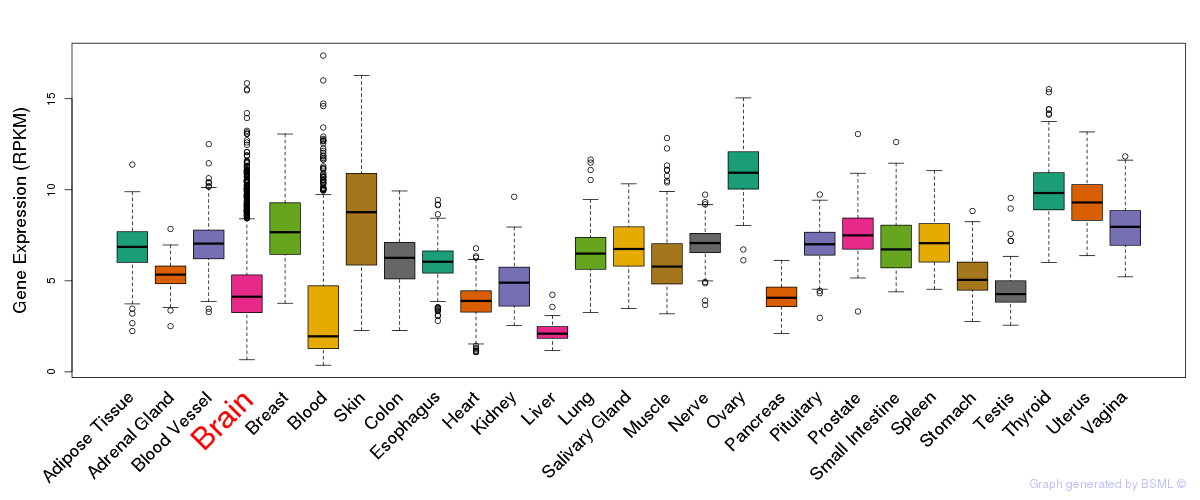
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003713 | transcription coactivator activity | NAS | 11734557 | |
| GO:0005515 | protein binding | IPI | 12200431 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0016568 | chromatin modification | IEA | - | |
| GO:0048096 | chromatin-mediated maintenance of transcription | NAS | 11734557 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016514 | SWI/SNF complex | IDA | 11734557 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 6 | 52 | 32 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 1370 | 1377 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-129-5p | 952 | 958 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC | ||||
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-142-5p | 1320 | 1326 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-25/32/92/363/367 | 489 | 496 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 1371 | 1377 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-329 | 194 | 200 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-342 | 22 | 28 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-34b | 760 | 767 | 1A,m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-377 | 21 | 27 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-382 | 1177 | 1183 | m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-410 | 922 | 928 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-433-3p | 1070 | 1076 | 1A | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-493-5p | 1004 | 1011 | 1A,m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU | ||||
| miR-496 | 1264 | 1270 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 241 | 247 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.