Gene Page: PCDH19
Summary ?
| GeneID | 57526 |
| Symbol | PCDH19 |
| Synonyms | EFMR|EIEE9 |
| Description | protocadherin 19 |
| Reference | MIM:300460|HGNC:HGNC:14270|Ensembl:ENSG00000165194|HPRD:11843|Vega:OTTHUMG00000022000 |
| Gene type | protein-coding |
| Map location | Xq22.1 |
| Sherlock p-value | 0.87 |
| Fetal beta | -0.28 |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
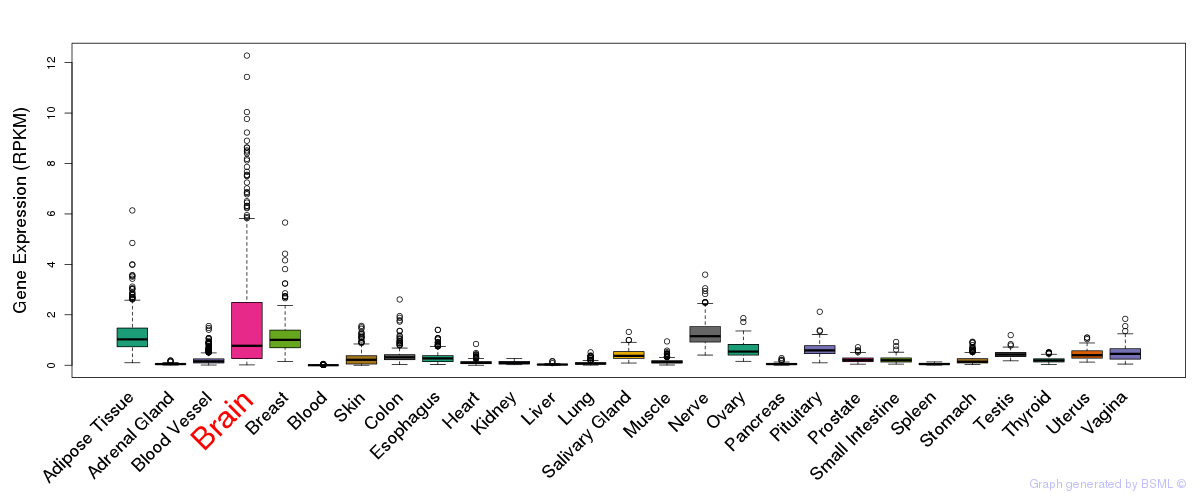
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADNP | 0.96 | 0.93 |
| ZNF445 | 0.96 | 0.92 |
| TUG1 | 0.95 | 0.94 |
| USP24 | 0.95 | 0.95 |
| HP1BP3 | 0.95 | 0.93 |
| TBC1D14 | 0.95 | 0.94 |
| ASXL1 | 0.95 | 0.92 |
| CLASP1 | 0.95 | 0.93 |
| TCEB3 | 0.95 | 0.92 |
| C6orf174 | 0.95 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.74 | -0.74 |
| HLA-F | -0.72 | -0.72 |
| AF347015.27 | -0.72 | -0.83 |
| AF347015.31 | -0.72 | -0.84 |
| MT-CO2 | -0.71 | -0.85 |
| S100B | -0.71 | -0.77 |
| AIFM3 | -0.70 | -0.70 |
| FXYD1 | -0.70 | -0.81 |
| CA4 | -0.70 | -0.73 |
| IFI27 | -0.70 | -0.82 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 DN | 39 | 21 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |