Gene Page: KLHL1
Summary ?
| GeneID | 57626 |
| Symbol | KLHL1 |
| Synonyms | MRP2 |
| Description | kelch like family member 1 |
| Reference | MIM:605332|HGNC:HGNC:6352|Ensembl:ENSG00000150361|HPRD:05623|Vega:OTTHUMG00000017056 |
| Gene type | protein-coding |
| Map location | 13q21 |
| Pascal p-value | 0.003 |
| Fetal beta | 3.697 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12917718 | 13 | 70682019 | KLHL1;ATXN8OS | 9.84E-5 | -0.214 | 0.028 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
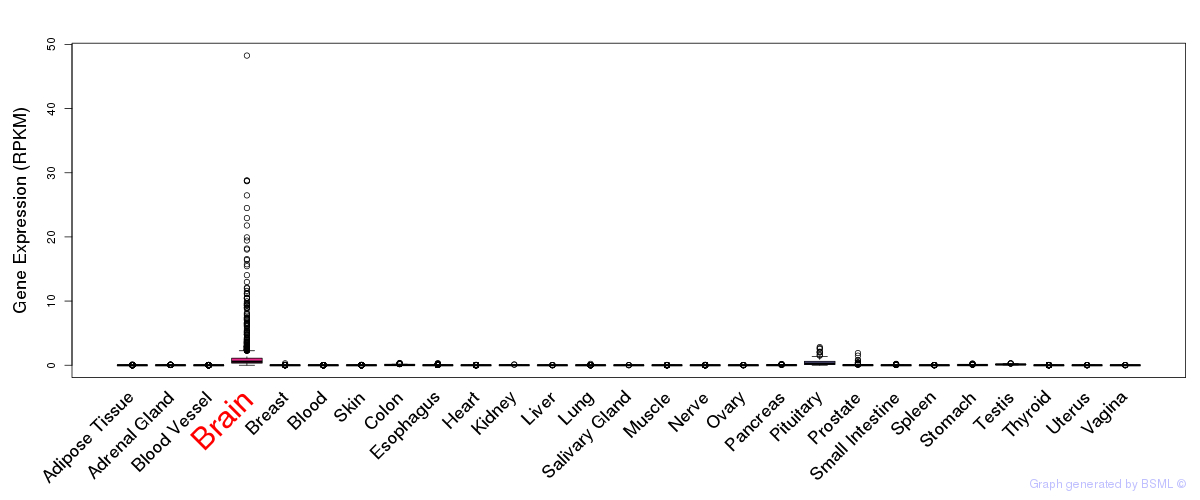
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | NAS | 10888605 | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0021680 | cerebellar Purkinje cell layer development | IEA | neuron, axon, Synap, dendrite (GO term level: 12) | - |
| GO:0016358 | dendrite development | IEA | neurite, dendrite (GO term level: 11) | - |
| GO:0007628 | adult walking behavior | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0030036 | actin cytoskeleton organization | NAS | 10888605 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | IEA | axon, dendrite (GO term level: 4) | - |
| GO:0030425 | dendrite | IEA | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | NAS | 10888605 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS | 82 | 55 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-24 | 727 | 734 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-452 | 669 | 675 | 1A | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-496 | 1061 | 1068 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-9 | 732 | 738 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.