Gene Page: PTMS
Summary ?
| GeneID | 5763 |
| Symbol | PTMS |
| Synonyms | ParaT |
| Description | parathymosin |
| Reference | MIM:168440|HGNC:HGNC:9629|Ensembl:ENSG00000159335|HPRD:01344|Vega:OTTHUMG00000169196 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.041 |
| Sherlock p-value | 0.182 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16854451 | chr3 | 108039512 | PTMS | 5763 | 0.16 | trans |
Section II. Transcriptome annotation
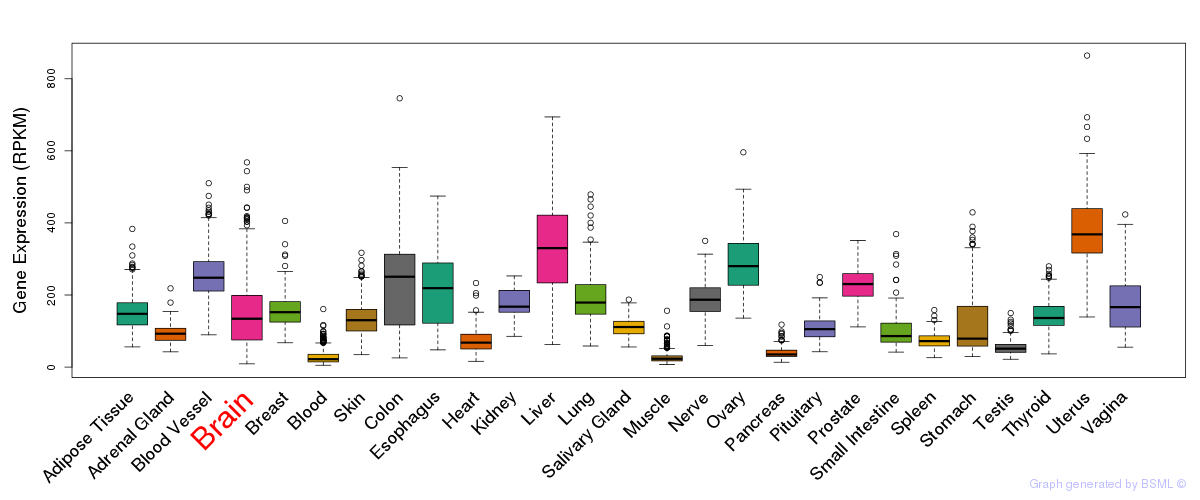
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TGOLN2 | 0.92 | 0.94 |
| PHLPP2 | 0.91 | 0.92 |
| TTPAL | 0.90 | 0.91 |
| SYT11 | 0.90 | 0.91 |
| USP32 | 0.90 | 0.93 |
| ATP2A2 | 0.89 | 0.91 |
| ENTPD4 | 0.89 | 0.92 |
| KCND3 | 0.88 | 0.93 |
| GABBR2 | 0.88 | 0.84 |
| TLN2 | 0.88 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.50 | -0.50 |
| C1orf61 | -0.48 | -0.58 |
| C1orf54 | -0.47 | -0.56 |
| GNG11 | -0.46 | -0.50 |
| HIGD1B | -0.44 | -0.47 |
| VAMP5 | -0.44 | -0.44 |
| AP002478.3 | -0.44 | -0.47 |
| DBI | -0.44 | -0.52 |
| SYCP3 | -0.43 | -0.51 |
| PLA2G5 | -0.43 | -0.42 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KORKOLA TERATOMA UP | 16 | 11 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE DN | 103 | 67 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM AND RAPAMYCIN SENSITIVE GENES | 68 | 35 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION | 37 | 20 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |