Gene Page: PTPN7
Summary ?
| GeneID | 5778 |
| Symbol | PTPN7 |
| Synonyms | BPTP-4|HEPTP|LC-PTP|LPTP|PTPNI |
| Description | protein tyrosine phosphatase, non-receptor type 7 |
| Reference | MIM:176889|HGNC:HGNC:9659|Ensembl:ENSG00000143851|HPRD:01480|Vega:OTTHUMG00000035931 |
| Gene type | protein-coding |
| Map location | 1q32.1 |
| Fetal beta | -0.326 |
| eGene | Cerebellum Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16869851 | chr4 | 20690056 | PTPN7 | 5778 | 0.09 | trans |
Section II. Transcriptome annotation
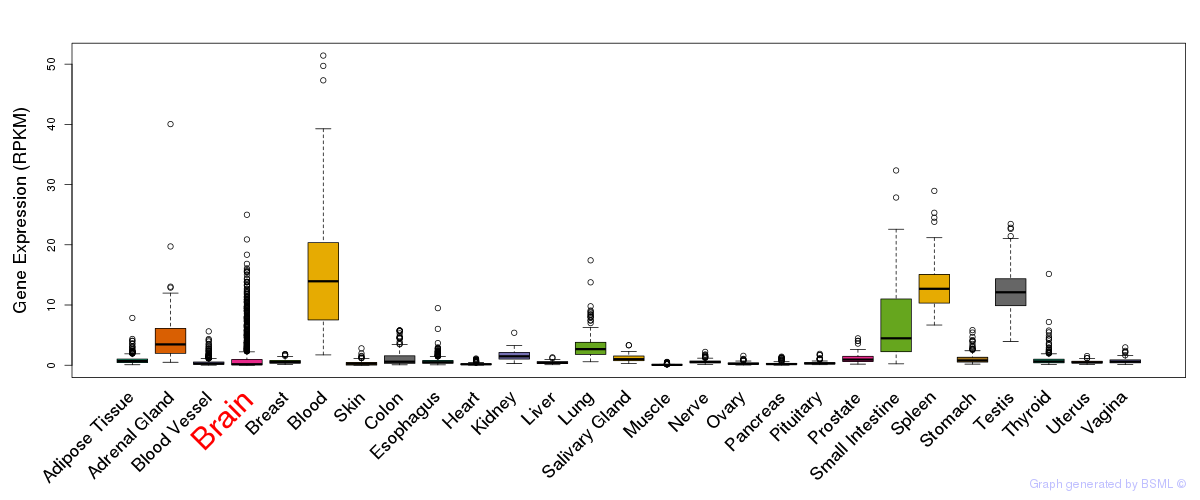
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC006116.3 | 0.90 | 0.91 |
| KPNA4 | 0.90 | 0.90 |
| CYB5R4 | 0.90 | 0.90 |
| HSPA14 | 0.89 | 0.89 |
| IPO5 | 0.89 | 0.89 |
| VPS37A | 0.89 | 0.88 |
| FAF1 | 0.89 | 0.87 |
| MRPS27 | 0.88 | 0.89 |
| ARL2BP | 0.88 | 0.88 |
| AC087742.2 | 0.88 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.78 | -0.81 |
| MT-CYB | -0.77 | -0.79 |
| MT-CO2 | -0.77 | -0.78 |
| AF347015.15 | -0.76 | -0.79 |
| AF347015.26 | -0.76 | -0.78 |
| AF347015.31 | -0.75 | -0.77 |
| AF347015.2 | -0.75 | -0.76 |
| AF347015.33 | -0.75 | -0.77 |
| AF347015.27 | -0.75 | -0.77 |
| AF347015.21 | -0.71 | -0.76 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| ADDYA ERYTHROID DIFFERENTIATION BY HEMIN | 73 | 47 | All SZGR 2.0 genes in this pathway |
| JAIN NFKB SIGNALING | 75 | 44 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |