Gene Page: CCNB1IP1
Summary ?
| GeneID | 57820 |
| Symbol | CCNB1IP1 |
| Synonyms | C14orf18|HEI10 |
| Description | cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| Reference | MIM:608249|HGNC:HGNC:19437|Ensembl:ENSG00000100814|HPRD:07000|Vega:OTTHUMG00000029509 |
| Gene type | protein-coding |
| Map location | 14q11.2 |
| Pascal p-value | 0.897 |
| Sherlock p-value | 0.86 |
| Fetal beta | 1.291 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2318860 | chr14 | 20743268 | CCNB1IP1 | 57820 | 3.849E-5 | cis | ||
| rs10134743 | chr14 | 20761615 | CCNB1IP1 | 57820 | 0.04 | cis | ||
| rs11623837 | chr14 | 20762065 | CCNB1IP1 | 57820 | 0 | cis | ||
| rs7160770 | chr14 | 20802706 | CCNB1IP1 | 57820 | 0.02 | cis | ||
| rs2318860 | chr14 | 20743268 | CCNB1IP1 | 57820 | 0 | trans | ||
| rs11623837 | chr14 | 20762065 | CCNB1IP1 | 57820 | 0.2 | trans | ||
| rs12589751 | 14 | 20747195 | CCNB1IP1 | ENSG00000100814.13 | 1.743E-7 | 0 | 54276 | gtex_brain_ba24 |
| rs11623318 | 14 | 20748396 | CCNB1IP1 | ENSG00000100814.13 | 3.446E-7 | 0 | 53075 | gtex_brain_ba24 |
| rs71410222 | 14 | 20752105 | CCNB1IP1 | ENSG00000100814.13 | 1.536E-7 | 0 | 49366 | gtex_brain_ba24 |
| rs536234118 | 14 | 20753577 | CCNB1IP1 | ENSG00000100814.13 | 2.07E-7 | 0 | 47894 | gtex_brain_ba24 |
| rs12888889 | 14 | 20753667 | CCNB1IP1 | ENSG00000100814.13 | 4.72E-7 | 0 | 47804 | gtex_brain_ba24 |
| rs1057605 | 14 | 20755287 | CCNB1IP1 | ENSG00000100814.13 | 2.27E-7 | 0 | 46184 | gtex_brain_ba24 |
| rs1953549 | 14 | 20760066 | CCNB1IP1 | ENSG00000100814.13 | 1.712E-8 | 0 | 41405 | gtex_brain_ba24 |
| rs55971623 | 14 | 20760432 | CCNB1IP1 | ENSG00000100814.13 | 4.348E-8 | 0 | 41039 | gtex_brain_ba24 |
| rs1000608 | 14 | 20763006 | CCNB1IP1 | ENSG00000100814.13 | 1.713E-8 | 0 | 38465 | gtex_brain_ba24 |
| rs1953552 | 14 | 20764585 | CCNB1IP1 | ENSG00000100814.13 | 1.441E-8 | 0 | 36886 | gtex_brain_ba24 |
| rs12890140 | 14 | 20764806 | CCNB1IP1 | ENSG00000100814.13 | 1.441E-8 | 0 | 36665 | gtex_brain_ba24 |
| rs2001572 | 14 | 20767868 | CCNB1IP1 | ENSG00000100814.13 | 2.174E-8 | 0 | 33603 | gtex_brain_ba24 |
| rs11845291 | 14 | 20777424 | CCNB1IP1 | ENSG00000100814.13 | 2.374E-9 | 0 | 24047 | gtex_brain_ba24 |
| rs11848535 | 14 | 20777431 | CCNB1IP1 | ENSG00000100814.13 | 2.028E-9 | 0 | 24040 | gtex_brain_ba24 |
| rs12588213 | 14 | 20779176 | CCNB1IP1 | ENSG00000100814.13 | 5.217E-9 | 0 | 22295 | gtex_brain_ba24 |
| rs3784244 | 14 | 20780367 | CCNB1IP1 | ENSG00000100814.13 | 6.229E-9 | 0 | 21104 | gtex_brain_ba24 |
| rs4451865 | 14 | 20783357 | CCNB1IP1 | ENSG00000100814.13 | 5.11E-9 | 0 | 18114 | gtex_brain_ba24 |
| rs12889262 | 14 | 20784117 | CCNB1IP1 | ENSG00000100814.13 | 5.11E-9 | 0 | 17354 | gtex_brain_ba24 |
| rs1132644 | 14 | 20784718 | CCNB1IP1 | ENSG00000100814.13 | 5.11E-9 | 0 | 16753 | gtex_brain_ba24 |
| rs7142395 | 14 | 20785289 | CCNB1IP1 | ENSG00000100814.13 | 8.918E-8 | 0 | 16182 | gtex_brain_ba24 |
| rs28500547 | 14 | 20785555 | CCNB1IP1 | ENSG00000100814.13 | 2.098E-7 | 0 | 15916 | gtex_brain_ba24 |
| rs6575132 | 14 | 20787779 | CCNB1IP1 | ENSG00000100814.13 | 2.193E-7 | 0 | 13692 | gtex_brain_ba24 |
| rs201930194 | 14 | 20790779 | CCNB1IP1 | ENSG00000100814.13 | 8.258E-8 | 0 | 10692 | gtex_brain_ba24 |
| rs1889361 | 14 | 20797836 | CCNB1IP1 | ENSG00000100814.13 | 2.972E-7 | 0 | 3635 | gtex_brain_ba24 |
| rs71108594 | 14 | 20800558 | CCNB1IP1 | ENSG00000100814.13 | 3.744E-7 | 0 | 913 | gtex_brain_ba24 |
| rs7160770 | 14 | 20802707 | CCNB1IP1 | ENSG00000100814.13 | 3.169E-7 | 0 | -1236 | gtex_brain_ba24 |
| rs4981975 | 14 | 20807898 | CCNB1IP1 | ENSG00000100814.13 | 3.797E-7 | 0 | -6427 | gtex_brain_ba24 |
| rs10625250 | 14 | 20819908 | CCNB1IP1 | ENSG00000100814.13 | 9.574E-7 | 0 | -18437 | gtex_brain_ba24 |
| rs10134743 | 14 | 20761616 | CCNB1IP1 | ENSG00000100814.13 | 1.851E-6 | 0.01 | 39855 | gtex_brain_putamen_basal |
| rs11623837 | 14 | 20762066 | CCNB1IP1 | ENSG00000100814.13 | 1.704E-6 | 0.01 | 39405 | gtex_brain_putamen_basal |
| rs10147548 | 14 | 20762775 | CCNB1IP1 | ENSG00000100814.13 | 1.506E-6 | 0.01 | 38696 | gtex_brain_putamen_basal |
| rs11845291 | 14 | 20777424 | CCNB1IP1 | ENSG00000100814.13 | 1.766E-6 | 0.01 | 24047 | gtex_brain_putamen_basal |
| rs11848535 | 14 | 20777431 | CCNB1IP1 | ENSG00000100814.13 | 1.935E-6 | 0.01 | 24040 | gtex_brain_putamen_basal |
| rs12588213 | 14 | 20779176 | CCNB1IP1 | ENSG00000100814.13 | 1.865E-6 | 0.01 | 22295 | gtex_brain_putamen_basal |
| rs7142395 | 14 | 20785289 | CCNB1IP1 | ENSG00000100814.13 | 8.128E-7 | 0.01 | 16182 | gtex_brain_putamen_basal |
| rs6575132 | 14 | 20787779 | CCNB1IP1 | ENSG00000100814.13 | 1.226E-6 | 0.01 | 13692 | gtex_brain_putamen_basal |
| rs201930194 | 14 | 20790779 | CCNB1IP1 | ENSG00000100814.13 | 3.817E-7 | 0.01 | 10692 | gtex_brain_putamen_basal |
| rs6575164 | 14 | 20792329 | CCNB1IP1 | ENSG00000100814.13 | 1.961E-6 | 0.01 | 9142 | gtex_brain_putamen_basal |
| rs1889361 | 14 | 20797836 | CCNB1IP1 | ENSG00000100814.13 | 7.312E-7 | 0.01 | 3635 | gtex_brain_putamen_basal |
| rs71108594 | 14 | 20800558 | CCNB1IP1 | ENSG00000100814.13 | 3.78E-7 | 0.01 | 913 | gtex_brain_putamen_basal |
| rs7160770 | 14 | 20802707 | CCNB1IP1 | ENSG00000100814.13 | 4.76E-7 | 0.01 | -1236 | gtex_brain_putamen_basal |
| rs4981975 | 14 | 20807898 | CCNB1IP1 | ENSG00000100814.13 | 3.455E-7 | 0.01 | -6427 | gtex_brain_putamen_basal |
| rs10625250 | 14 | 20819908 | CCNB1IP1 | ENSG00000100814.13 | 5.328E-7 | 0.01 | -18437 | gtex_brain_putamen_basal |
| rs1713419 | 14 | 20834548 | CCNB1IP1 | ENSG00000100814.13 | 6.239E-7 | 0.01 | -33077 | gtex_brain_putamen_basal |
| rs1713418 | 14 | 20834809 | CCNB1IP1 | ENSG00000100814.13 | 2.771E-7 | 0.01 | -33338 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
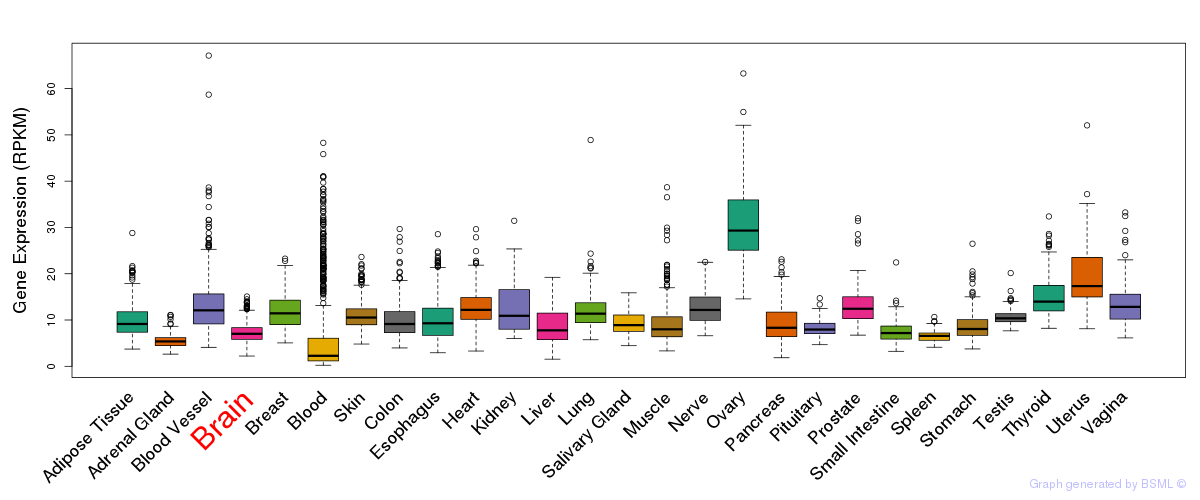
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12612082 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| ZERBINI RESPONSE TO SULINDAC DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG DN | 85 | 56 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE UP | 79 | 40 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |