Gene Page: SLAMF7
Summary ?
| GeneID | 57823 |
| Symbol | SLAMF7 |
| Synonyms | 19A|CD319|CRACC|CS1 |
| Description | SLAM family member 7 |
| Reference | MIM:606625|HGNC:HGNC:21394|Ensembl:ENSG00000026751|HPRD:09432|Vega:OTTHUMG00000024008 |
| Gene type | protein-coding |
| Map location | 1q23.1-q24.1 |
| Fetal beta | -0.227 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
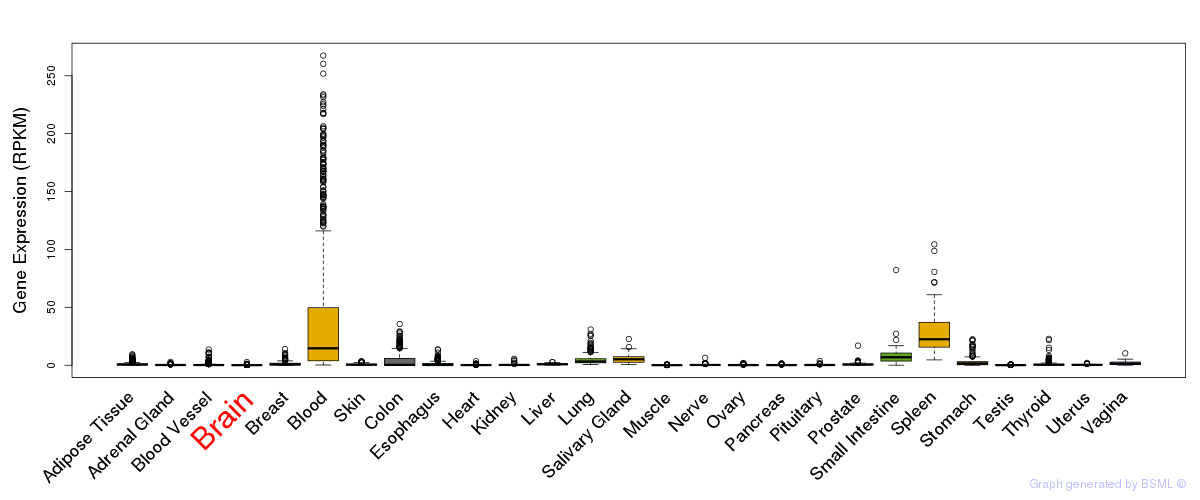
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030101 | natural killer cell activation | NAS | 11698418 | |
| GO:0007155 | cell adhesion | NAS | 11802771 | |
| GO:0042267 | natural killer cell mediated cytotoxicity | NAS | 11698418 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | NAS | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 MULTIPLE MYELOMA PROGRAM | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE | 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED DENDRITIC CELL | 65 | 49 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |