Gene Page: PTPRB
Summary ?
| GeneID | 5787 |
| Symbol | PTPRB |
| Synonyms | HPTP-BETA|HPTPB|PTPB|R-PTP-BETA|VEPTP |
| Description | protein tyrosine phosphatase, receptor type B |
| Reference | MIM:176882|HGNC:HGNC:9665|Ensembl:ENSG00000127329|HPRD:01474|Vega:OTTHUMG00000169499 |
| Gene type | protein-coding |
| Map location | 12q15 |
| Pascal p-value | 0.124 |
| Fetal beta | -1.167 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2498500 | chr1 | 119673092 | PTPRB | 5787 | 0.04 | trans | ||
| rs17033855 | chr2 | 107766740 | PTPRB | 5787 | 0.16 | trans | ||
| rs2096481 | chr8 | 36570335 | PTPRB | 5787 | 0.1 | trans | ||
| rs16885322 | chr8 | 36598931 | PTPRB | 5787 | 0.1 | trans | ||
| rs10510107 | chr10 | 124029470 | PTPRB | 5787 | 0.09 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
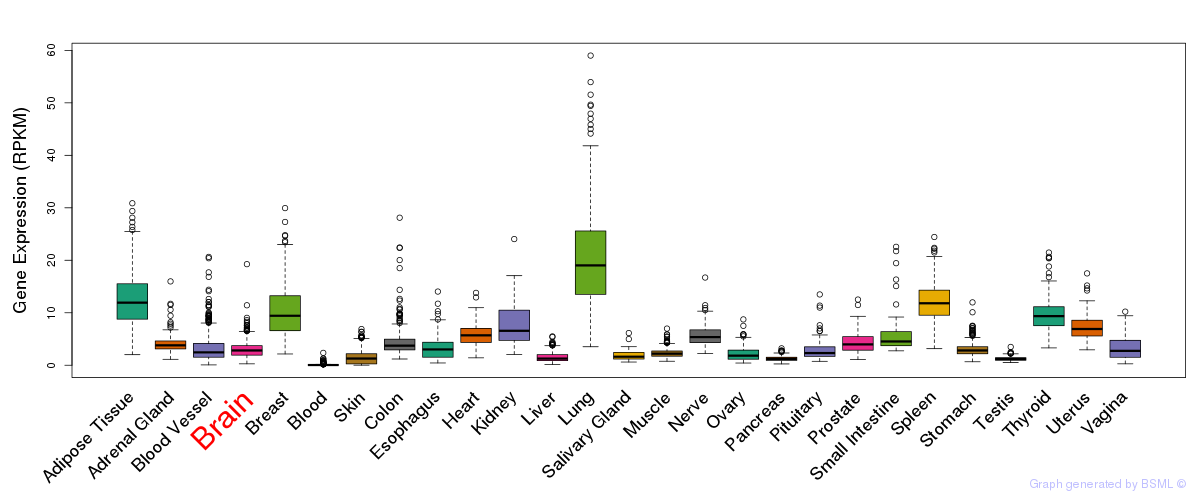
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CPNE2 | 0.82 | 0.79 |
| GPR172A | 0.81 | 0.76 |
| C19orf63 | 0.81 | 0.77 |
| MMP25 | 0.80 | 0.76 |
| ISYNA1 | 0.79 | 0.80 |
| AC100793.2 | 0.79 | 0.80 |
| SPNS1 | 0.79 | 0.76 |
| GNB2 | 0.79 | 0.72 |
| PCDHGC4 | 0.78 | 0.80 |
| LTB4R2 | 0.78 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.58 | -0.60 |
| ACOT13 | -0.54 | -0.60 |
| FAM162A | -0.54 | -0.60 |
| KBTBD3 | -0.52 | -0.62 |
| CA4 | -0.52 | -0.51 |
| ASPA | -0.52 | -0.62 |
| AF347015.27 | -0.52 | -0.56 |
| SLCO1A2 | -0.51 | -0.60 |
| EVI2A | -0.51 | -0.67 |
| OMG | -0.51 | -0.55 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPRY PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST PRENEOPLASTIC DN | 55 | 33 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN | 149 | 93 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 12Q13 Q21 AMPLICON | 46 | 21 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |