Gene Page: PYGM
Summary ?
| GeneID | 5837 |
| Symbol | PYGM |
| Synonyms | - |
| Description | phosphorylase, glycogen, muscle |
| Reference | MIM:608455|HGNC:HGNC:9726|Ensembl:ENSG00000068976|HPRD:01986|Vega:OTTHUMG00000066835 |
| Gene type | protein-coding |
| Map location | 11q12-q13.2 |
| Pascal p-value | 0.265 |
| Sherlock p-value | 0.083 |
| Fetal beta | -1 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09958483 | 11 | 64527240 | PYGM | 5.93E-4 | 0.37 | 0.05 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11256073 | chr10 | 9152267 | PYGM | 5837 | 0.09 | trans | ||
| rs10509155 | chr10 | 63401917 | PYGM | 5837 | 0.08 | trans |
Section II. Transcriptome annotation
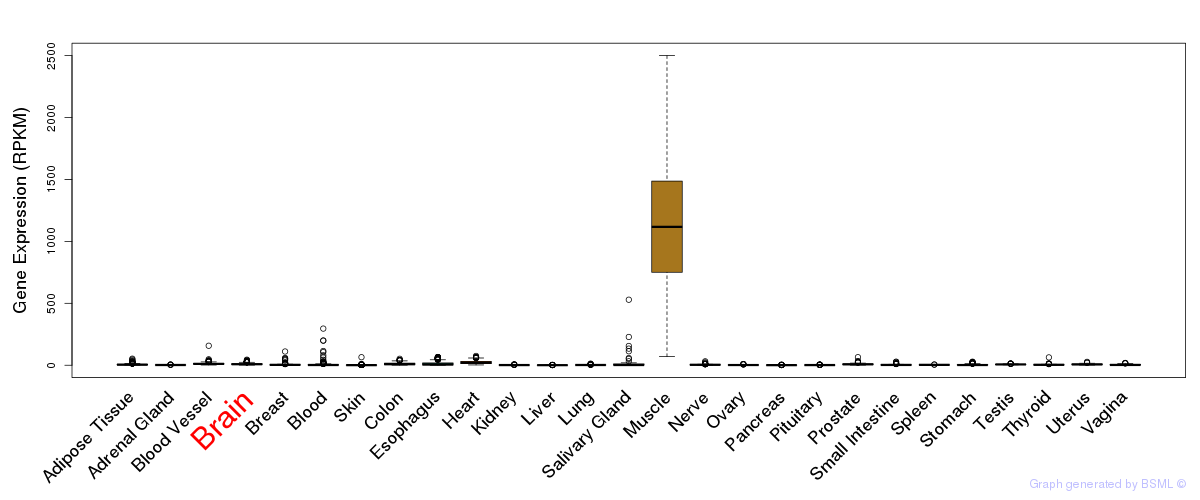
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MFN2 | 0.89 | 0.87 |
| BCL2L13 | 0.88 | 0.86 |
| PTK2 | 0.88 | 0.84 |
| SYT11 | 0.87 | 0.89 |
| KIAA1737 | 0.87 | 0.85 |
| LONP2 | 0.87 | 0.84 |
| RAB7A | 0.87 | 0.81 |
| NPLOC4 | 0.86 | 0.83 |
| FBXW11 | 0.86 | 0.83 |
| WIPI2 | 0.86 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.67 | -0.46 |
| IL32 | -0.62 | -0.48 |
| GNG11 | -0.58 | -0.43 |
| AF347015.18 | -0.57 | -0.39 |
| AC098691.2 | -0.57 | -0.46 |
| C1orf54 | -0.55 | -0.46 |
| FAM159B | -0.55 | -0.50 |
| CLEC3B | -0.54 | -0.50 |
| SYCP3 | -0.54 | -0.48 |
| C16orf74 | -0.54 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STARCH AND SUCROSE METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FEEDER PATHWAY | 9 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | 18 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| KUNINGER IGF1 VS PDGFB TARGETS UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOGEN METABOLISM | 21 | 16 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |