Gene Page: RAC1
Summary ?
| GeneID | 5879 |
| Symbol | RAC1 |
| Synonyms | MIG5|Rac-1|TC-25|p21-Rac1 |
| Description | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| Reference | MIM:602048|HGNC:HGNC:9801|Ensembl:ENSG00000136238|HPRD:03627|Vega:OTTHUMG00000023540 |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 0.705 |
| Sherlock p-value | 0.56 |
| Fetal beta | -0.426 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2217 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15381475 | 7 | 6436101 | RAC1 | 1.34E-4 | 0.506 | 0.031 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
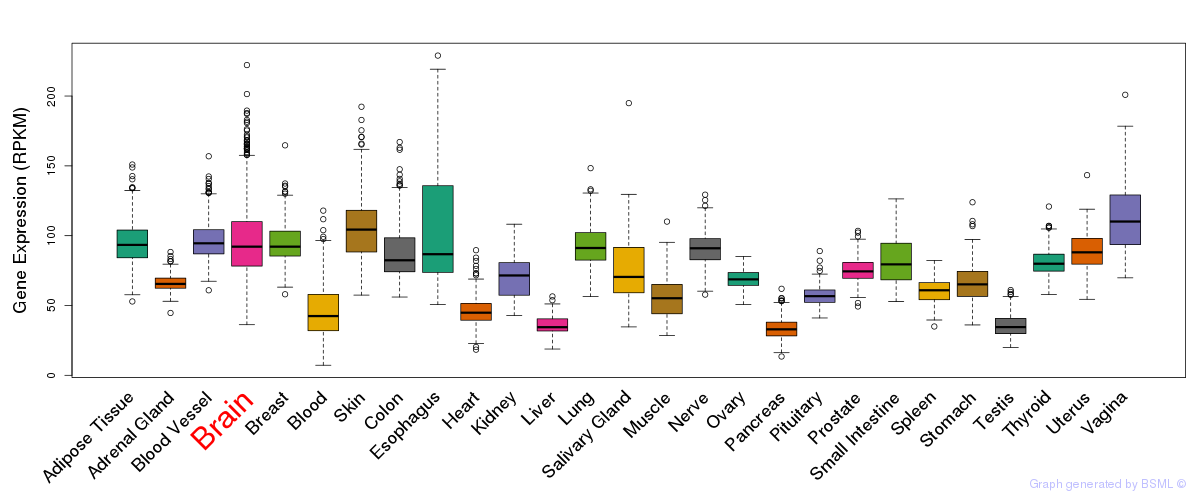
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPIRE1 | 0.91 | 0.95 |
| SLAIN2 | 0.90 | 0.91 |
| NELL2 | 0.89 | 0.88 |
| RLBP1L1 | 0.89 | 0.89 |
| NCOA2 | 0.89 | 0.94 |
| ACVR1B | 0.89 | 0.93 |
| LASS6 | 0.88 | 0.93 |
| KBTBD7 | 0.88 | 0.93 |
| HERC3 | 0.88 | 0.92 |
| ZNF238 | 0.88 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.67 | -0.84 |
| SERPINB6 | -0.65 | -0.72 |
| HSD17B14 | -0.64 | -0.76 |
| MT-CO2 | -0.64 | -0.82 |
| AF347015.31 | -0.64 | -0.80 |
| S100B | -0.62 | -0.77 |
| AF347015.33 | -0.62 | -0.77 |
| TSC22D4 | -0.62 | -0.73 |
| HEPN1 | -0.62 | -0.71 |
| AIFM3 | -0.62 | -0.71 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 9312003 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0019899 | enzyme binding | IPI | 16636067 | |
| GO:0030742 | GTP-dependent protein binding | IPI | 10954424 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 9572733 | |
| GO:0009653 | anatomical structure morphogenesis | TAS | 9572733 | |
| GO:0010310 | regulation of hydrogen peroxide metabolic process | TAS | 16636067 | |
| GO:0006954 | inflammatory response | TAS | 9572733 | |
| GO:0006928 | cell motion | TAS | 9572733 | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0035025 | positive regulation of Rho protein signal transduction | TAS | 9312003 | |
| GO:0031529 | ruffle organization | TAS | 9312003 | |
| GO:0048261 | negative regulation of receptor-mediated endocytosis | TAS | 9312003 | |
| GO:0030041 | actin filament polymerization | TAS | 9312003 | |
| GO:0030032 | lamellipodium biogenesis | IMP | 9312003 | |
| GO:0051668 | localization within membrane | IMP | 16636067 | |
| GO:0060263 | regulation of respiratory burst | IDA | 16636067 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 7744773 |10648409 |14737186 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTA1 | ACTA | ASMA | CFTD | CFTD1 | CFTDM | MPFD | NEM1 | NEM2 | NEM3 | actin, alpha 1, skeletal muscle | - | HPRD | 11027608 |
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD,BioGRID | 11027608 |
| ARFIP2 | POR1 | ADP-ribosylation factor interacting protein 2 | Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 8670882 |11346801 |11478794 |
| ARFIP2 | POR1 | ADP-ribosylation factor interacting protein 2 | - | HPRD | 8670882 |11478794 |
| ARHGAP15 | BM046 | Rho GTPase activating protein 15 | - | HPRD,BioGRID | 12650940 |
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Biochemical Activity | BioGRID | 11431473 |
| ARHGAP27 | CAMGAP1 | FLJ43547 | MGC120624 | Rho GTPase activating protein 27 | Biochemical Activity | BioGRID | 15147912 |
| ARHGAP8 | BPGAP1 | FLJ20185 | PP610 | Rho GTPase activating protein 8 | BPGAP1 interacts with Rac1. | BIND | 12944407 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | - | HPRD | 10346909 |10673424|11513579 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | - | HPRD,BioGRID | 10673424 |
| ARHGDIB | D4 | GDIA2 | GDID4 | LYGDI | Ly-GDI | RAP1GN1 | RhoGDI2 | Rho GDP dissociation inhibitor (GDI) beta | - | HPRD,BioGRID | 9799233 |
| ARHGDIG | RHOGDI-3 | Rho GDP dissociation inhibitor (GDI) gamma | Rac1 interacts with RhoGDI. | BIND | 14764880 |
| ARHGEF2 | DKFZp547L106 | DKFZp547P1516 | GEF | GEF-H1 | GEFH1 | KIAA0651 | LFP40 | P40 | rho/rac guanine nucleotide exchange factor (GEF) 2 | - | HPRD,BioGRID | 9857026 |11595749 |
| ARHGEF4 | ASEF | ASEF1 | GEF4 | STM6 | Rho guanine nucleotide exchange factor (GEF) 4 | Reconstituted Complex | BioGRID | 10947987 |
| ARHGEF6 | COOL2 | Cool-2 | KIAA0006 | MRX46 | PIXA | alpha-PIX | alphaPIX | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | ARHGEF6 (Cool-2) interacts with Rac1. | BIND | 15649357 |
| BAIAP2 | BAP2 | IRSP53 | BAI1-associated protein 2 | - | HPRD,BioGRID | 11130076 |12054568 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | Two-hybrid | BioGRID | 9427749 |
| CDC42BPG | DMPK2 | HSMDPKIN | KAPPA-200 | MRCKgamma | CDC42 binding protein kinase gamma (DMPK-like) | - | HPRD,BioGRID | 15194684 |
| CDC42SE1 | SCIP1 | SPEC1 | CDC42 small effector 1 | - | HPRD,BioGRID | 10816584 |
| CDC42SE2 | FLJ21967 | SPEC2 | CDC42 small effector 2 | Two-hybrid | BioGRID | 10816584 |
| CDGAP | KIAA1204 | MGC138368 | MGC138370 | Cdc42 GTPase-activating protein | - | HPRD | 9786927 |
| CHN1 | ARHGAP2 | CHN | DURS2 | RHOGAP2 | chimerin (chimaerin) 1 | - | HPRD | 8496137 |
| CIT | CRIK | KIAA0949 | STK21 | citron (rho-interacting, serine/threonine kinase 21) | Far Western | BioGRID | 8543060 |
| CLIP1 | CLIP | CLIP-170 | CLIP170 | CYLN1 | MGC131604 | RSN | CAP-GLY domain containing linker protein 1 | Affinity Capture-Western | BioGRID | 12110184 |
| CYBA | p22-PHOX | cytochrome b-245, alpha polypeptide | - | HPRD | 10486263 |
| CYFIP1 | FLJ45151 | P140SRA-1 | SHYC | SRA1 | cytoplasmic FMR1 interacting protein 1 | - | HPRD | 9417078 |
| DEF6 | IBP | SLAT | differentially expressed in FDCP 6 homolog (mouse) | - | HPRD,BioGRID | 15023524 |
| DEF6 | IBP | SLAT | differentially expressed in FDCP 6 homolog (mouse) | DEF6 interacts with Rac1. This interaction was modelled on a demonstrated interaction between human DEF6 and Rac1 from an unspecified species. | BIND | 15023524 |
| DIAPH1 | DFNA1 | DIA1 | DRF1 | FLJ25265 | LFHL1 | hDIA1 | diaphanous homolog 1 (Drosophila) | - | HPRD | 11590143 |
| DMPK | DM | DM1 | DM1PK | DMK | MDPK | MT-PK | dystrophia myotonica-protein kinase | Reconstituted Complex | BioGRID | 10869570 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | Rac interacts with Dock180. | BIND | 12134158 |
| DOCK1 | DOCK180 | ced5 | dedicator of cytokinesis 1 | - | HPRD,BioGRID | 9808620 |
| DOCK2 | FLJ46592 | KIAA0209 | dedicator of cytokinesis 2 | - | HPRD,BioGRID | 10559471 |
| DOCK8 | FLJ00026 | FLJ00152 | FLJ00346 | MRD2 | ZIR8 | dedicator of cytokinesis 8 | - | HPRD,BioGRID | 15304341 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD | 7738010 |
| FHOD1 | FHOS | formin homology 2 domain containing 1 | - | HPRD,BioGRID | 11590143 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 10051605 |
| FMNL1 | C17orf1 | C17orf1B | FHOD4 | FMNL | KW-13 | MGC133052 | MGC1894 | MGC21878 | formin-like 1 | - | HPRD,BioGRID | 10958683 |
| GEFT | p63RhoGEF | RhoA/RAC/CDC42 exchange factor | - | HPRD,BioGRID | 12547822 |
| ICMT | HSTE14 | MGC39955 | MST098 | MSTP098 | PCCMT | PCMT | PPMT | isoprenylcysteine carboxyl methyltransferase | - | HPRD,BioGRID | 12006387 |
| IL1RAP | C3orf13 | FLJ37788 | IL-1RAcP | IL1R3 | interleukin 1 receptor accessory protein | - | HPRD,BioGRID | 11416133 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | - | HPRD,BioGRID | 8670801 |
| IQGAP2 | - | IQ motif containing GTPase activating protein 2 | - | HPRD,BioGRID | 8702968 |8756646|8756646 |
| IQGAP2 | - | IQ motif containing GTPase activating protein 2 | - | HPRD | 8756646 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | - | HPRD,BioGRID | 8769096 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | Reconstituted Complex | BioGRID | 9427749 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | - | HPRD | 9427749 |
| MAP3K4 | FLJ42439 | KIAA0213 | MAPKKK4 | MEKK4 | MTK1 | PRO0412 | mitogen-activated protein kinase kinase kinase 4 | Reconstituted Complex | BioGRID | 9079650 |
| MCF2L | ARHGEF14 | DBS | FLJ12122 | KIAA0362 | OST | MCF.2 cell line derived transforming sequence-like | - | HPRD,BioGRID | 7957046 |14701795 |
| METAP2 | MAP2 | MNPEP | p67 | p67eIF2 | methionyl aminopeptidase 2 | - | HPRD | 7738010 |
| MYD88 | MYD88D | myeloid differentiation primary response gene (88) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11416133 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | - | HPRD | 9255350 |
| NCF2 | FLJ93058 | NCF-2 | NOXA2 | P67-PHOX | P67PHOX | neutrophil cytosolic factor 2 | - | HPRD | 9255350 |9624165 |11896062|11090627 |
| NCF2 | FLJ93058 | NCF-2 | NOXA2 | P67-PHOX | P67PHOX | neutrophil cytosolic factor 2 | - | HPRD | 7738010 |8036496 |9255350 |9624165 |11090627 |11896062|11090627 |
| NCF2 | FLJ93058 | NCF-2 | NOXA2 | P67-PHOX | P67PHOX | neutrophil cytosolic factor 2 | Co-crystal Structure Reconstituted Complex | BioGRID | 9624165 |9642115 |11090627 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 10766742 |
| NCKAP1 | FLJ11291 | HEM2 | KIAA0587 | MGC8981 | NAP1 | NAP125 | NCK-associated protein 1 | - | HPRD | 9148763 |9417078 |
| NCKAP1 | FLJ11291 | HEM2 | KIAA0587 | MGC8981 | NAP1 | NAP125 | NCK-associated protein 1 | - | HPRD,BioGRID | 9148763 |
| NOS2 | HEP-NOS | INOS | NOS | NOS2A | nitric oxide synthase 2, inducible | - | HPRD | 11422783 |11457725 |
| NOS2 | HEP-NOS | INOS | NOS | NOS2A | nitric oxide synthase 2, inducible | - | HPRD,BioGRID | 11457725 |
| NOXA1 | FLJ25475 | MGC131800 | NY-CO-31 | SDCCAG31 | p51NOX | NADPH oxidase activator 1 | - | HPRD | 12716910 |
| OPHN1 | MRX60 | OPN1 | oligophrenin 1 | - | HPRD,BioGRID | 9582072 |11998687 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | PAK1 interacts with an unspecified isoform of Rac. This interaction was modeled on a demonstrated interaction between human PAK1 and Rac from Chinese hamster. | BIND | 15975516 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | PAK1 interacts with an unspecified isoform of Rac. This interaction was modeled on a demonstrated interaction between PAK1 from an unspecified species and human Rac. | BIND | 15864311 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | - | HPRD | 7744004 |8107774 |11134022 |11950930|11134022 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Reconstituted Complex | BioGRID | 12650940 |12879077 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Active GTP-bound RAC1 interacts with PAK1B. This interaction was modelled on a demonstrated interaction between chicken RAC1 and human PAK1B. | BIND | 15735674 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Rac1 interacts with Pak1. This interaction was modeled on a demonstrated interaction between Rac1 and Pak1 from an unspecified species. | BIND | 15652748 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Rac1 interacts with Pak1. This interaction was modelled on a demonstrated interacton between Rac1 from an unspecified species and human Pak1. | BIND | 12944407 |
| PAK2 | PAK65 | PAKgamma | p21 protein (Cdc42/Rac)-activated kinase 2 | - | HPRD,BioGRID | 9535855 |
| PAK3 | CDKN1A | MRX30 | MRX47 | OPHN3 | PAK3beta | bPAK | hPAK3 | p21 protein (Cdc42/Rac)-activated kinase 3 | Reconstituted Complex Two-hybrid | BioGRID | 11478794 |
| PAK4 | - | p21 protein (Cdc42/Rac)-activated kinase 4 | Affinity Capture-Western | BioGRID | 11756552 |
| PAK7 | KIAA1264 | MGC26232 | PAK5 | p21 protein (Cdc42/Rac)-activated kinase 7 | - | HPRD,BioGRID | 11756552 |
| PARD6A | PAR-6A | PAR6 | PAR6C | PAR6alpha | TAX40 | TIP-40 | par-6 partitioning defective 6 homolog alpha (C. elegans) | - | HPRD,BioGRID | 11260256 |
| PARD6G | FLJ45701 | PAR-6G | PAR6gamma | par-6 partitioning defective 6 homolog gamma (C. elegans) | - | HPRD,BioGRID | 11260256 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | Two-hybrid | BioGRID | 16169070 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 7629060 |8034624 |12086876 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 12086876 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD | 11950936 |
| PLEKHG2 | CLG | DKFZp667J2325 | FLJ00018 | FLJ22458 | FLJ38638 | pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | - | HPRD,BioGRID | 11839748 |
| PLXNB1 | KIAA0407 | MGC149167 | PLEXIN-B1 | PLXN5 | SEP | plexin B1 | - | HPRD | 12123608 |
| PLXNB1 | KIAA0407 | MGC149167 | PLEXIN-B1 | PLXN5 | SEP | plexin B1 | Plexin-B1 interacts with Rac1. This interaction was modelled on a demonstrated interaction between human Plexin-B1 and Rac1 from an unspecified species. | BIND | 15642257 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 11284700 |
| PTPLAD1 | B-IND1 | FLJ90376 | HSPC121 | protein tyrosine phosphatase-like A domain containing 1 | - | HPRD,BioGRID | 10747961 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Reconstituted Complex | BioGRID | 11134022 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | Rac1 interacts with RLIP76. | BIND | 7673236 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | - | HPRD,BioGRID | 7673236 |
| RAP1GDS1 | GDS1 | MGC118859 | MGC118861 | SmgGDS | RAP1, GTP-GDP dissociation stimulator 1 | - | HPRD,BioGRID | 11948427 |
| RCC2 | DKFZp762N0610 | KIAA1470 | TD-60 | regulator of chromosome condensation 2 | - | HPRD | 12919680 |
| RGL2 | HKE1.5 | KE1.5 | RAB2L | ral guanine nucleotide dissociation stimulator-like 2 | Two-hybrid | BioGRID | 8939933 |
| RICH2 | KIAA0672 | NPC-A-10 | Rho-type GTPase-activating protein RICH2 | Biochemical Activity | BioGRID | 11431473 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | Biochemical Activity | BioGRID | 12454018 |12819203 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | - | HPRD | 11438723 |
| SH3BP1 | - | SH3-domain binding protein 1 | - | HPRD | 10508610 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 11021801 |
| SYNJ2 | INPP5H | KIAA0348 | MGC44422 | synaptojanin 2 | - | HPRD,BioGRID | 11084340 |
| TBC1D3 | TBC1D3A | TRE17 | TBC1 domain family, member 3 | - | HPRD,BioGRID | 12612085 |
| TCF7L2 | TCF-4 | TCF4 | transcription factor 7-like 2 (T-cell specific, HMG-box) | Rac1 interacts with TCF4. | BIND | 15377999 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | Phenotypic Enhancement | BioGRID | 11328862 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | The GTPase Rac1 specifically interacts with Dbl-homology domain of the nucleotide exchange factor TIAM1. This interaction is modelled on a demonstrated interaction between mouse TIAM1 and human Rac1. | BIND | 11130063 |
| TIAM1 | FLJ36302 | T-cell lymphoma invasion and metastasis 1 | - | HPRD,BioGRID | 11130063 |11595749 |
| TNFRSF12A | CD266 | FN14 | TWEAKR | tumor necrosis factor receptor superfamily, member 12A | - | HPRD | 14573547 |
| TNFRSF12A | CD266 | FN14 | TWEAKR | tumor necrosis factor receptor superfamily, member 12A | Fn14 interacts with Rac1. | BIND | 14573547 |
| TRIO | FLJ42780 | tgat | triple functional domain (PTPRF interacting) | - | HPRD,BioGRID | 11595749 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | - | HPRD | 8631991 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 11287617 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10523675 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD,BioGRID | 8643625 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | - | HPRD | 9843499 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY | 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SALMONELLA PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PYK2 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P38MAPK PATHWAY | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EDG1 PATHWAY | 27 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAC1 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKCELLS PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAL PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDC42RAC PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARF PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACTINY PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID IL2 PI3K PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID NEPHRIN NEPH1 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| PID AR NONGENOMIC PATHWAY | 31 | 27 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID RAC1 REG PATHWAY | 38 | 25 | All SZGR 2.0 genes in this pathway |
| PID ARF 3PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID EPHA2 FWD PATHWAY | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID RAC1 PATHWAY | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF RAC | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT VAV1 PATHWAY | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ROBO RECEPTOR | 30 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME DSCAM INTERACTIONS | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL UP | 73 | 50 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIMOFEEVA GROWTH STRESS VIA STAT1 DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS REVERTED UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA DELAYED RESPONSE TO TGFB1 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C4 | 13 | 10 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL IL3RA | 9 | 6 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 537 | 544 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-137 | 987 | 993 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-142-3p | 747 | 754 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-144 | 538 | 544 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-182 | 308 | 315 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-194 | 127 | 133 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-200bc/429 | 715 | 721 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-361 | 1018 | 1025 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-485-3p | 1493 | 1499 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-485-5p | 466 | 472 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-495 | 125 | 131 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-96 | 309 | 315 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| miR-99/100 | 777 | 783 | 1A | hsa-miR-99abrain | AACCCGUAGAUCCGAUCUUGUG |
| hsa-miR-100brain | AACCCGUAGAUCCGAACUUGUG | ||||
| hsa-miR-99bbrain | CACCCGUAGAACCGACCUUGCG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.