Gene Page: RAP1A
Summary ?
| GeneID | 5906 |
| Symbol | RAP1A |
| Synonyms | C21KG|G-22K|KREV-1|KREV1|RAP1|SMGP21 |
| Description | RAP1A, member of RAS oncogene family |
| Reference | MIM:179520|HGNC:HGNC:9855|Ensembl:ENSG00000116473|HPRD:01545|Vega:OTTHUMG00000011959 |
| Gene type | protein-coding |
| Map location | 1p13.3 |
| Pascal p-value | 0.058 |
| Sherlock p-value | 0.359 |
| Fetal beta | -0.359 |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12303540 | chr12 | 132609678 | RAP1A | 5906 | 0.16 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
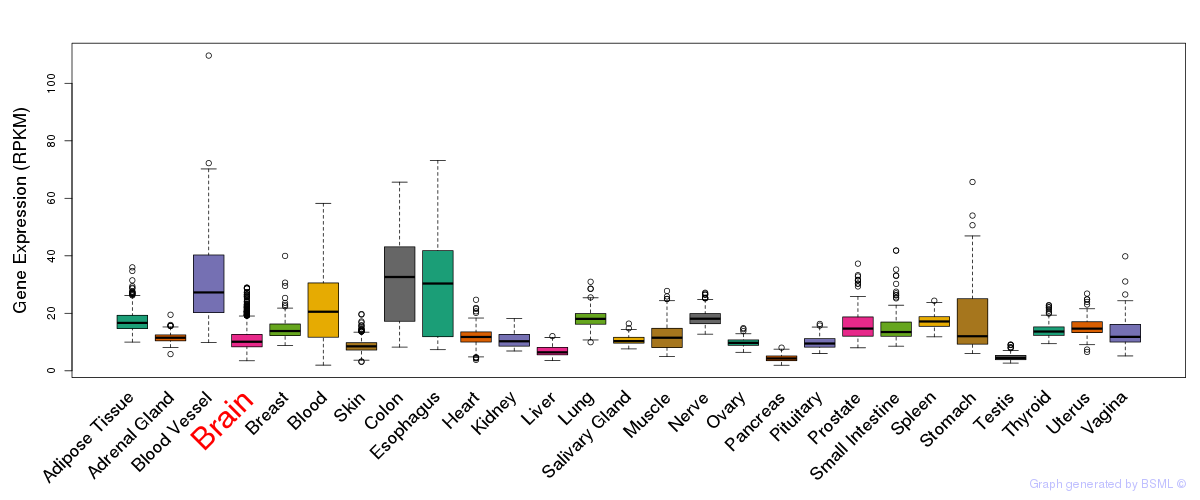
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 10777492 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11022048 | |
| GO:0005525 | GTP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 16284401 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBB1IP | INAG1 | PREL1 | RARP1 | RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | - | HPRD | 15469846 |
| APBB1IP | INAG1 | PREL1 | RARP1 | RIAM | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | RIAM interacts with Rap1. | BIND | 15469846 |
| ARHGEF1 | GEF1 | LBCL2 | LSC | P115-RHOGEF | SUB1.5 | Rho guanine nucleotide exchange factor (GEF) 1 | Biochemical Activity | BioGRID | 12581858 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | Reconstituted Complex | BioGRID | 10454553 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 15854902 |
| GABARAPL2 | ATG8 | GATE-16 | GATE16 | GEF-2 | GEF2 | GABA(A) receptor-associated protein-like 2 | Biochemical Activity | BioGRID | 12581858 |
| KRIT1 | CAM | CCM1 | KRIT1, ankyrin repeat containing | - | HPRD,BioGRID | 9285558 |
| MLLT4 | AF-6 | AF6 | AFADIN | FLJ34371 | RP3-431P23.3 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | - | HPRD,BioGRID | 10224125 |10922060|10922060 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | Affinity Capture-Western | BioGRID | 11466412 |
| PDE6D | PDED | phosphodiesterase 6D, cGMP-specific, rod, delta | - | HPRD,BioGRID | 11786539 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | - | HPRD | 11335720 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 7791872 |7862125 |9867809 |10454553 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD | 7791872|9867809|10454553 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | Two-hybrid | BioGRID | 11786539 |
| RALGDS | FLJ20922 | RGF | RalGEF | ral guanine nucleotide dissociation stimulator | - | HPRD,BioGRID | 10085114 |
| RALGDS | FLJ20922 | RGF | RalGEF | ral guanine nucleotide dissociation stimulator | Rap1A interacts with RalGDS. This interaction was modeled on a demonstrated interaction between human Rap1A and RalGDS from an unspecified species. | BIND | 15856025 |
| RAP1GAP | KIAA0474 | RAP1GA1 | Rap1GAP1 | rap1GAPII | RAP1 GTPase activating protein | Biochemical Activity | BioGRID | 12842888 |
| RAP1GDS1 | GDS1 | MGC118859 | MGC118861 | SmgGDS | RAP1, GTP-GDP dissociation stimulator 1 | - | HPRD,BioGRID | 11948427 |
| RAPGEF1 | C3G | DKFZp781P1719 | GRF2 | Rap guanine nucleotide exchange factor (GEF) 1 | Rap1A interacts with an unspecified isoform of C3G. | BIND | 15856025 |
| RAPGEF2 | CNrasGEF | NRAPGEP | PDZ-GEF1 | PDZGEF1 | RA-GEF | Rap-GEP | Rap guanine nucleotide exchange factor (GEF) 2 | Biochemical Activity Reconstituted Complex | BioGRID | 10934204 |
| RAPGEF3 | CAMP-GEFI | EPAC | EPAC1 | HSU79275 | MGC21410 | bcm910 | Rap guanine nucleotide exchange factor (GEF) 3 | Biochemical Activity | BioGRID | 10777494 |
| RAPGEF3 | CAMP-GEFI | EPAC | EPAC1 | HSU79275 | MGC21410 | bcm910 | Rap guanine nucleotide exchange factor (GEF) 3 | Rap1A interacts with Epac1. | BIND | 15856025 |
| RAPGEF4 | CAMP-GEFII | CGEF2 | EPAC2 | Nbla00496 | Rap guanine nucleotide exchange factor (GEF) 4 | Biochemical Activity | BioGRID | 10777494 |
| RAPGEF4 | CAMP-GEFII | CGEF2 | EPAC2 | Nbla00496 | Rap guanine nucleotide exchange factor (GEF) 4 | Rap1A interacts with Epac2. This interaction was modeled on a demonstrated interaction between human Rap1A and mouse Epac2. | BIND | 15856025 |
| RAPGEF5 | GFR | KIAA0277 | MR-GEF | REPAC | Rap guanine nucleotide exchange factor (GEF) 5 | Rap1A interacts with Repac. | BIND | 15856025 |
| RAPGEF5 | GFR | KIAA0277 | MR-GEF | REPAC | Rap guanine nucleotide exchange factor (GEF) 5 | Biochemical Activity | BioGRID | 10777494 |10934204 |
| RAPGEF6 | DKFZp667N084 | DKFZp686I15116 | KIA001LB | PDZ-GEF2 | PDZGEF2 | RA-GEF-2 | Rap guanine nucleotide exchange factor (GEF) 6 | - | HPRD,BioGRID | 11524421 |12581858 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | in vivo | BioGRID | 2164710 |
| RASGRP4 | - | RAS guanyl releasing protein 4 | - | HPRD | 11880369 |
| RASIP1 | FLJ20401 | RAIN | Ras interacting protein 1 | - | HPRD | 15031288 |
| RGL4 | MGC119678 | MGC119680 | Rgr | ral guanine nucleotide dissociation stimulator-like 4 | - | HPRD,BioGRID | 12874025 |
| RGS14 | - | regulator of G-protein signaling 14 | - | HPRD,BioGRID | 10926822 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | Affinity Capture-Western | BioGRID | 15854902 |
| RUNDC3A | RAP2IP | RPIP8 | RUN domain containing 3A | Reconstituted Complex | BioGRID | 9523700 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western Co-purification | BioGRID | 12147258 |12842888 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID RET PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME ARMS MEDIATED ACTIVATION | 17 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLONGED ERK ACTIVATION EVENTS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | 15 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME RAP1 SIGNALLING | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D DN | 31 | 19 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS DN | 24 | 12 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 72 | 78 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-149 | 23 | 30 | 1A,m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-19 | 154 | 161 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-203.1 | 299 | 305 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-23 | 127 | 133 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24 | 21 | 28 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-25/32/92/363/367 | 229 | 235 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 260 | 266 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-299-5p | 628 | 634 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-320 | 677 | 683 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-324-5p | 294 | 300 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-330 | 544 | 550 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-410 | 118 | 124 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-433-3p | 355 | 361 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-539 | 219 | 225 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-543 | 306 | 312 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.